Integrated barcode chips for rapid, multiplexed analysis of proteins in microliter quantities of blood
- PMID: 19029914
- PMCID: PMC2775523
- DOI: 10.1038/nbt.1507
Integrated barcode chips for rapid, multiplexed analysis of proteins in microliter quantities of blood
Abstract
As the tissue that contains the largest representation of the human proteome, blood is the most important fluid for clinical diagnostics. However, although changes of plasma protein profiles reflect physiological or pathological conditions associated with many human diseases, only a handful of plasma proteins are routinely used in clinical tests. Reasons for this include the intrinsic complexity of the plasma proteome, the heterogeneity of human diseases and the rapid degradation of proteins in sampled blood. We report an integrated microfluidic system, the integrated blood barcode chip that can sensitively sample a large panel of protein biomarkers over broad concentration ranges and within 10 min of sample collection. It enables on-chip blood separation and rapid measurement of a panel of plasma proteins from quantities of whole blood as small as those obtained by a finger prick. Our device holds potential for inexpensive, noninvasive and informative clinical diagnoses, particularly in point-of-care settings.
Figures

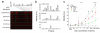
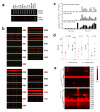

Comment in
-
Microfluidics closes in on point-of-care assays.Nat Biotechnol. 2008 Dec;26(12):1345-6. doi: 10.1038/nbt1208-1345. Nat Biotechnol. 2008. PMID: 19060870 No abstract available.
References
-
- Anderson NL, Anderson NG. The human plasma proteome - History, character, and diagnostic prospects. Molecular & Cellular Proteomics. 2002;1:845–867. - PubMed
-
- Fujii K, et al. Clinical-scale high-throughput human plasma proteome clinical analysis: Lung adenocarcinoma. Proteomics. 2005;5:1150–1159. - PubMed
-
- Lathrop JT, Anderson NL, Anderson NG, Hammond DJ. Therapeutic potential of the plasma proteome. Current Opinion in Molecular Therapeutics. 2003;5:250–257. - PubMed
-
- Hsieh SY, Chen RK, Pan YH, Lee HL. Systematical evaluation of the effects of sample collection procedures on low-molecular-weight serum/plasma proteome profiling. Proteomics. 2006;6:3189–3198. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

