Dlx1&2 and Mash1 transcription factors control striatal patterning and differentiation through parallel and overlapping pathways
- PMID: 19030180
- PMCID: PMC2761428
- DOI: 10.1002/cne.21854
Dlx1&2 and Mash1 transcription factors control striatal patterning and differentiation through parallel and overlapping pathways
Abstract
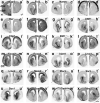
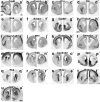
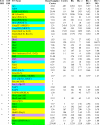
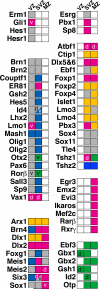
Here we define the expression of approximately 100 transcription factors in progenitors and neurons of the developing basal ganglia. We have begun to elucidate the transcriptional hierarchy of these genes with respect to the Dlx homeodomain genes, which are essential for differentiation of most GABAergic projection neurons of the basal ganglia. This analysis identified Dlx-dependent and Dlx-independent pathways. The Dlx-independent pathway depends in part on the function of the Mash1 b-HLH transcription factor. These analyses define core transcriptional components that differentially specify the identity and differentiation of the striatum, nucleus accumbens, and septum.
2008 Wiley-Liss, Inc.
Figures






References
-
- Anderson S, Mione M, Yun K, Rubenstein JL. Differential origins of neocortical projection and local circuit neurons: role of Dlx genes in neocortical interneuronogenesis. Cereb Cortex. 1999;9(6):646–654. - PubMed
-
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997a;278(5337):474–476. - PubMed
-
- Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JL. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron. 1997b;19(1):27–37. - PubMed
-
- Bulfone A, Kim HJ, Puelles L, Porteus MH, Grippo JF, Rubenstein JL. The mouse Dlx-2 (Tes-1) gene is expressed in spatially restricted domains of the forebrain, face and limbs in midgestation mouse embryos. Mech Dev. 1993a;40(3):129–140. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

