The chemistry and biochemistry of heme c: functional bases for covalent attachment
- PMID: 19030605
- PMCID: PMC2654777
- DOI: 10.1039/b717196j
The chemistry and biochemistry of heme c: functional bases for covalent attachment
Abstract
A discussion of the literature concerning the synthesis, function, and activity of heme c-containing proteins is presented. Comparison of the properties of heme c, which is covalently bound to protein, is made to heme b, which is bound noncovalently. A question of interest is why nature uses biochemically expensive heme c in many proteins when its properties are expected to be similar to heme b. Considering the effects of covalent heme attachment on heme conformation and on the proximal histidine interaction with iron, it is proposed that heme attachment influences both heme reduction potential and ligand-iron interactions.
Figures




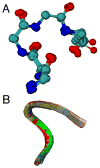
References
-
- Chapman SK, Daff S, Munro AW. Metal Sites in Proteins and Models. 1997:39–70.
-
- Schneider S, Marles-Wright J, Sharp KH, Paoli M. Nat Prod Rep. 2007;24:621–630. - PubMed
-
- Paoli M, Marles-Wright J, Smith A. DNA Cell Biol. 2002;21:271–280. - PubMed
-
- Stevens JM, Daltrop O, Allen JWA, Ferguson SJ. Acc Chem Res. 2004;37:999–1007. - PubMed
-
- Barker PD, Ferguson SJ. Structure. 1999;7:R281–R290. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

