Non-solid oncogenes in solid tumors: EML4-ALK fusion genes in lung cancer
- PMID: 19032370
- PMCID: PMC11158085
- DOI: 10.1111/j.1349-7006.2008.00972.x
Non-solid oncogenes in solid tumors: EML4-ALK fusion genes in lung cancer
Abstract
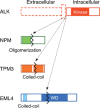
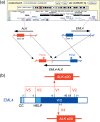
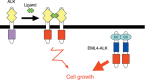
It is generally accepted that recurrent chromosome translocations play a major role in the molecular pathogenesis of hematological malignancies but not of solid tumors. However, chromosome translocations involving the e26 transformation-specific sequence transcription factor loci have been demonstrated recently in many prostate cancer cases. Furthermore, through a functional screening with retroviral cDNA expression libraries, we have discovered the fusion-type protein tyrosine kinase echinoderm microtubule-associated protein like-4 (EML4)-anaplastic lymphoma kinase (ALK) in non-small cell lung cancer (NSCLC) specimens. A recurrent chromosome translocation, inv(2)(p21p23), in NSCLC generates fused mRNA encoding the amino-terminal half of EML4 ligated to the intracellular region of the receptor-type protein tyrosine kinase ALK. EML4-ALK oligomerizes constitutively in cells through the coiled coil domain within the EML4 region, and becomes activated to exert a marked oncogenicity both in vitro and in vivo. Break and fusion points within the EML4 locus may diverge in NSCLC cells to generate various isoforms of EML4-ALK, which may constitute approximately 5% of NSCLC cases, at least in the Asian ethnic group. In the present review I summarize how detection of EML4-ALK cDNA may become a sensitive diagnostic means for NSCLC cases that are positive for the fusion gene, and discuss whether suppression of ALK enzymatic activity could be an effective treatment strategy against this intractable disorder.
Figures


References
-
- Ren R. Mechanisms of BCR‐ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer 2005; 5: 172–83. - PubMed
-
- Pear WS, Miller JP, Xu L et al . Efficient and rapid induction of a chronic myelogenous leukemia‐like myeloproliferative disease in mice receiving P210 bcr/abl‐transduced bone marrow. Blood 1998; 92: 3780–92. - PubMed
-
- Oehler VG, Radich JP. Monitoring bcr‐abl by polymerase chain reaction in the treatment of chronic myeloid leukemia. Curr Oncol Rep 2003; 5: 426–35. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

