New insights into the origin of the B genome of hexaploid wheat: evolutionary relationships at the SPA genomic region with the S genome of the diploid relative Aegilops speltoides
- PMID: 19032732
- PMCID: PMC2612700
- DOI: 10.1186/1471-2164-9-555
New insights into the origin of the B genome of hexaploid wheat: evolutionary relationships at the SPA genomic region with the S genome of the diploid relative Aegilops speltoides
Abstract
Background: Several studies suggested that the diploid ancestor of the B genome of tetraploid and hexaploid wheat species belongs to the Sitopsis section, having Aegilops speltoides (SS, 2n = 14) as the closest identified relative. However molecular relationships based on genomic sequence comparison, including both coding and non-coding DNA, have never been investigated. In an attempt to clarify these relationships, we compared, in this study, sequences of the Storage Protein Activator (SPA) locus region of the S genome of Ae. speltoides (2n = 14) to that of the A, B and D genomes co-resident in the hexaploid wheat species (Triticum aestivum, AABBDD, 2n = 42).
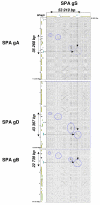
Results: Four BAC clones, spanning the SPA locus of respectively the A, B, D and S genomes, were isolated and sequenced. Orthologous genomic regions were identified as delimited by shared non-transposable elements and non-coding sequences surrounding the SPA gene and correspond to 35,268, 22,739, 43,397 and 53,919 bp for the A, B, D and S genomes, respectively. Sequence length discrepancies within and outside the SPA orthologous regions are the result of non-shared transposable elements (TE) insertions, all of which inserted after the progenitors of the four genomes divergence.
Conclusion: On the basis of conserved sequence length as well as identity of the shared non-TE regions and the SPA coding sequence, Ae speltoides appears to be more evolutionary related to the B genome of T. aestivum than the A and D genomes. However, the differential insertions of TEs, none of which are conserved between the two genomes led to the conclusion that the S genome of Ae. speltoides has diverged very early from the progenitor of the B genome which remains to be identified.
Figures


References
-
- Gaut BS. Evolutionary dynamics of grass geno. New phytologist. 2002;154:15–28. doi: 10.1046/j.1469-8137.2002.00352.x. - DOI
-
- Harlan JR. Crops and Man. Madison, Wisconsin: American Society of Agronomy, Inc; 1992.
-
- Zohary D, Hopf M. Domestication of plants in the Old World. 3. New York: Oxford University Press; 2000. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

