Distribution of ancestral proto-Actinopterygian chromosome arms within the genomes of 4R-derivative salmonid fishes (Rainbow trout and Atlantic salmon)
- PMID: 19032764
- PMCID: PMC2632648
- DOI: 10.1186/1471-2164-9-557
Distribution of ancestral proto-Actinopterygian chromosome arms within the genomes of 4R-derivative salmonid fishes (Rainbow trout and Atlantic salmon)
Abstract
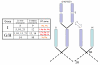
Background: Comparative genomic studies suggest that the modern day assemblage of ray-finned fishes have descended from an ancestral grouping of fishes that possessed 12-13 linkage groups. All jawed vertebrates are postulated to have experienced two whole genome duplications (WGD) in their ancestry (2R duplication). Salmonids have experienced one additional WGD (4R duplication event) compared to most extant teleosts which underwent a further 3R WGD compared to other vertebrates. We describe the organization of the 4R chromosomal segments of the proto-ray-finned fish karyotype in Atlantic salmon and rainbow trout based upon their comparative syntenies with two model species of 3R ray-finned fishes.
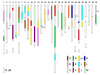
Results: Evidence is presented for the retention of large whole-arm affinities between the ancestral linkage groups of the ray-finned fishes, and the 50 homeologous chromosomal segments in Atlantic salmon and rainbow trout. In the comparisons between the two salmonid species, there is also evidence for the retention of large whole-arm homeologous affinities that are associated with the retention of duplicated markers. Five of the 7 pairs of chromosomal arm regions expressing the highest level of duplicate gene expression in rainbow trout share homologous synteny to the 5 pairs of homeologs with the greatest duplicate gene expression in Atlantic salmon. These regions are derived from proto-Actinopterygian linkage groups B, C, E, J and K.
Conclusion: Two chromosome arms in Danio rerio and Oryzias latipes (descendants of the 3R duplication) can, in most instances be related to at least 4 whole or partial chromosomal arms in the salmonid species. Multiple arm assignments in the two salmonid species do not clearly support a 13 proto-linkage group model, and suggest that a 12 proto-linkage group arrangement (i.e., a separate single chromosome duplication and ancestral fusion/fissions/recombination within the putative G/H/I groupings) may have occurred in the more basal soft-rayed fishes. We also found evidence supporting the model that ancestral linkage group M underwent a single chromosome duplication following the 3R duplication. In the salmonids, the M ancestral linkage groups are localized to 5 whole arm, and 3 partial arm regions (i.e., 6 whole arm regions expected). Thus, 3 distinct ancestral linkage groups are postulated to have existed in the G/H and M lineage chromosomes in the ancestor of the salmonids.
Figures




References
-
- Allendorf FW, Thorgaard GH. Tetraploidy and the evolution of salmonid fishes. In: Turner BJ, editor. Evolutionary Genetics of Fishes. Chap 1. Plenum Press, NY; 1984. pp. 1–46.
-
- Gharbi K, Gautier A, Danzmann RG, Gharbi S, Sakamoto T, Hoyheim B, Taggart JB, Cairney M, Powell R, Kreig F, et al. A linkage map for brown trout (Salmo trutta): Chromosome homeologies and comparative genome organization with other salmonid fish. Genetics. 2006;172:2405–2419. doi: 10.1534/genetics.105.048330. - DOI - PMC - PubMed
-
- Wright JE, Johnson K, Hollister A, May B. Meiotic models to explain classical linkage, pseudolinkage, and chromosome paring in tetraploid derivative salmonid genomes. Isozymes Curr Top Biol Med Res. 1983;10:239–260. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

