Inhibition of early endosome fusion by Rab5-binding defective Ras interference 1 mutants
- PMID: 19032933
- PMCID: PMC2761816
- DOI: 10.1016/j.abb.2008.11.009
Inhibition of early endosome fusion by Rab5-binding defective Ras interference 1 mutants
Abstract
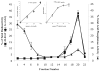
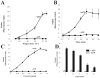
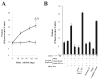
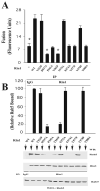
Rin1 has been shown to play an important role in endocytosis. In this study we demonstrated that depletion of Rin1 from the cytosol blocked the fusion reaction. More importantly, endosome fusion was rescued by the addition of Rin1 proteins depending on the presence of Rab5, and its effector EEA1. Furthermore, we found that Syntaxin 13, but not Syntaxin 7, was required by Rin1 to support endosome fusion. We also identified six mutations on the Vps9 domain of Rin1 that failed to rescue the fusion reaction. Two of them, Rin1: D537A and Rin1: Y561F mutants showed dramatic inhibitory effect on the fusion reaction, which correlate with their inability to properly activate Rab5 or to bind endosomal membranes. Taken together, our results suggest that specific residues on the Vsp9 domain of Rin1 are required for its interaction with Rab5, binding to the endosomal membranes and subsequent regulation of the fusion reaction.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

