Genetic changes accompanying the evolution of host specialization in Drosophila sechellia
- PMID: 19033155
- PMCID: PMC2644960
- DOI: 10.1534/genetics.108.093419
Genetic changes accompanying the evolution of host specialization in Drosophila sechellia
Abstract
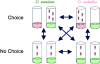
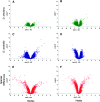
Changes in host specialization contribute to the diversification of phytophagous insects. When shifting to a new host, insects evolve new physiological, morphological, and behavioral adaptations. Our understanding of the genetic changes responsible for these adaptations is limited. For instance, we do not know how often host shifts involve gain-of-function vs. loss-of-function alleles. Recent work suggests that some genes involved in odor recognition are lost in specialists. Here we show that genes involved in detoxification and metabolism, as well as those affecting olfaction, have reduced gene expression in Drosophila sechellia-a specialist on the fruit of Morinda citrifolia. We screened for genes that differ in expression between D. sechellia and its generalist sister species, D. simulans. We also screened for genes that are differentially expressed in D. sechellia when these flies chose their preferred host vs. when they were forced onto other food. D. sechellia increases expression of genes involved with oogenesis and fatty acid metabolism when on its host. The majority of differentially expressed genes, however, appear downregulated in D. sechellia. For several functionally related genes, this decrease in expression is associated with apparent loss-of-function alleles. For example, the D. sechellia allele of Odorant binding protein 56e (Obp56e) harbors a premature stop codon. We show that knockdown of Obp56e activity significantly reduces the avoidance response of D. melanogaster toward M. citrifolia. We argue that apparent loss-of-function alleles like Obp56e potentially contributed to the initial adaptation of D. sechellia to its host. Our results suggest that a subset of genes reduce or lose function as a consequence of host specialization, which may explain why, in general, specialist insects tend to shift to chemically similar hosts.
Figures
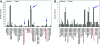

References
-
- Amlou, M., B. Moreteau and J. R. David, 1998. a Genetic analysis of Drosophila sechellia specialization: oviposition behavior toward the major aliphatic acids of its host plant. Behav. Genet. 28 455–464. - PubMed
-
- Amlou, M., B. Moreteau and J. R. David, 1998. b Larval tolerance in the Drosophila melanogaster species complex toward the two toxic acids of the D. sechellia host plant. Hereditas 129 7–14. - PubMed
-
- Anholt, R. R. H., and T. F. C. Mackay, 2001. The genetic architecture of odor-guided behavior in Drosophila melanogaster. Behav. Genet. 31 17–27. - PubMed
-
- Barker, J. S., W. T. Starmer and J. C. Fogleman, 1994. Genotype-specific habitat selection for oviposition sites in the cactophilic species Drosophila buzzatii. Heredity 72 384–395. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

