Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression
- PMID: 19033359
- PMCID: PMC2615625
- DOI: 10.1093/nar/gkn919
Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression
Abstract
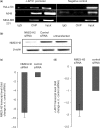
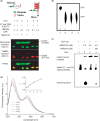
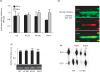
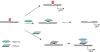
Regulatory influence of the G-quadruplex or G4 motif present within the nuclease hypersensitive element (NHE) in the promoter of c-MYC has been noted. On the other hand, association of NM23-H2 to the NHE leads to c-MYC activation. Therefore, NM23-H2 interaction with the G4 motif within the c-MYC NHE presents an interesting mechanistic possibility. Herein, using luciferase reporter assay and chromatin immunoprecipitation we show NM23-H2 mediated c-MYC activation involves NM23-H2-G4 motif binding within the c-MYC NHE. G4 motif complex formation with recombinant NM23-H2 was independently confirmed using fluorescence energy transfer, which also indicated that the G4 motif was resolved to an unfolded state within the protein-bound complex. Taken together, this supports transcriptional role of NM23-H2 via a G4 motif.
Figures

References
-
- Hatfield GW, Benham CJ. DNA topology-mediated control of global gene expression in Escherichia coli. Annu. Rev. Genet. 2002;36:175–203. - PubMed
-
- Bacolla A, Wells RD. Non-B DNA conformations, genomic rearrangements, and human disease. J. Biol. Chem. 2004;279:47411–47414. - PubMed
-
- Rich A, Zhang S. Timeline: Z-DNA: the long road to biological function. Nat. Rev. Genet. 2003;4:566–572. - PubMed
-
- Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. - PubMed
-
- Balagurumoorthy P, Brahmachari SK. Structure and stability of human telomeric sequence. J. Biol. Chem. 1994;269:21858–21869. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

