A large genome center's improvements to the Illumina sequencing system
- PMID: 19034268
- PMCID: PMC2610436
- DOI: 10.1038/nmeth.1270
A large genome center's improvements to the Illumina sequencing system
Abstract
The Wellcome Trust Sanger Institute is one of the world's largest genome centers, and a substantial amount of our sequencing is performed with 'next-generation' massively parallel sequencing technologies: in June 2008 the quantity of purity-filtered sequence data generated by our Genome Analyzer (Illumina) platforms reached 1 terabase, and our average weekly Illumina production output is currently 64 gigabases. Here we describe a set of improvements we have made to the standard Illumina protocols to make the library preparation more reliable in a high-throughput environment, to reduce bias, tighten insert size distribution and reliably obtain high yields of data.
Figures
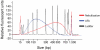
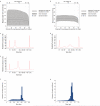
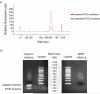
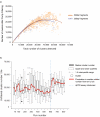
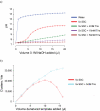
References
-
- Bentley DR. Whole-genome re-sequencing. Curr Opin Genet Dev. 2006;16:545–552. - PubMed
-
- Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. - PubMed
-
- Surzycki S. Basic Techniques in Molecular Biology. Springer-Verlag; Berlin: 2000. pp. 377–380.
-
- Albert TJ, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–905. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

