Measles virus entry inhibitors: a structural proposal for mechanism of action and the development of resistance
- PMID: 19035834
- PMCID: PMC4084900
- DOI: 10.1021/bi801513p
Measles virus entry inhibitors: a structural proposal for mechanism of action and the development of resistance
Abstract
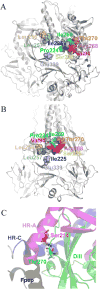
Previously, we developed a panel of nonpeptidic compounds specifically preventing fusion of the measles virus (MV) with target cells at IC(50) values of 0.6-3 muM. Mutations in the MV fusion protein (MV F) that render resistance to these blockers were described. The structural basis for both inhibition and resistance was unclear in the earlier work because of the availability of a structural model for only the postfusion conformation of MV F. We have now developed structural models for both pre- and postfusion conformers of the latter protein trimer. The models allow investigation of the large-scale conformational changes occurring in the MV fusion machinery and, in conjunction with antisera binding studies, provide a rationale for how inhibitors may arrest a conformational intermediate by interfering with the formation of interactions between the heptad repeat B (HR-B) linker and DIII domains. The models also show that resistance to inhibition can be explained by a predicted destabilizing effect of the mutations on the HR-B domain within the trimeric prefusion structure. This viewpoint is supported by the temperature-dependent differential fusion activities of MV F variants harboring these mutations.
Figures
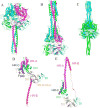






References
-
- World Health Organization. World Health Report, 2002. 2004. Annex 2.
-
- Colman PM, Lawrence MC. The structural biology of type I viral membrane fusion. Nat Rev Mol Cell Biol. 2003;4:309–319. - PubMed
-
- Kielian M, Jungerwirth S. Mechanisms of enveloped virus entry into cells. Mol Biol Med. 1990;7:17–31. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

