Biphasic decay kinetics suggest progressive slowing in turnover of latently HIV-1 infected cells during antiretroviral therapy
- PMID: 19036147
- PMCID: PMC2630982
- DOI: 10.1186/1742-4690-5-107
Biphasic decay kinetics suggest progressive slowing in turnover of latently HIV-1 infected cells during antiretroviral therapy
Abstract
Background: Mathematical models based on kinetics of HIV-1 plasma viremia after initiation of combination antiretroviral therapy (cART) inferred HIV-infected cells to decay exponentially with constant rates correlated to their strength of virus production. To further define in vivo decay kinetics of HIV-1 infected cells experimentally, we assessed infected cell-classes of distinct viral transcriptional activity in peripheral blood mononuclear cells (PBMC) of five patients during 1 year after initiation of cART RESULTS: In a novel analytical approach patient-matched PCR for unspliced and multiply spliced viral RNAs was combined with limiting dilution analysis at the single cell level. This revealed that HIV-RNA+ PBMC can be stratified into four distinct viral transcriptional classes. Two overlapping cell-classes of high viral transcriptional activity, suggestive of a virion producing phenotype, rapidly declined to undetectable levels. Two cell classes expressing HIV-RNA at low and intermediate levels, presumably insufficient for virus production and occurring at frequencies exceeding those of productively infected cells matched definitions of HIV-latency. These cells persisted during cART. Nevertheless, during the first four weeks of therapy their kinetics resembled that of productively infected cells.
Conclusion: We have observed biphasic decays of latently HIV-infected cells of low and intermediate viral transcriptional activity with marked decreases in cell numbers shortly after initiation of therapy and complete persistence in later phases. A similar decay pattern was shared by cells with greatly enhanced viral transcriptional activity which showed a certain grade of levelling off before their disappearance. Thus it is conceivable that turnover/decay rates of HIV-infected PBMC may be intrinsically variable. In particular they might be accelerated by HIV-induced activation and reactivation of the viral life cycle and slowed down by the disappearance of such feedback-loops after initiation of cART.
Figures
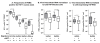





References
-
- Carpenter C, Feinberg M, Aubry W, Averitt D, Coffin J, Cooper D, Follansbee S, Hamburg P, Harrington M, Hidalgo J, et al. Report of the NIH Panel to Define Principles of Therapy of HIV Infection. Ann Intern Med. 1998;128:1057–1078. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

