Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers
- PMID: 19036716
- PMCID: PMC2724058
- DOI: 10.1194/jlr.D800051-JLR200
Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers
Abstract
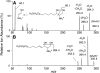
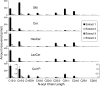
Sphingolipids are a highly diverse category of bioactive compounds. This article describes methods that have been validated for the extraction, liquid chromatographic (LC) separation, identification and quantitation of sphingolipids by electrospray ionization, tandem mass spectrometry (ESI-MS/MS) using triple quadrupole (QQQ, API 3000) and quadrupole-linear-ion trap (API 4000 QTrap, operating in QQQ mode) mass spectrometers. Advantages of the QTrap included: greater sensitivity, similar ionization efficiencies for sphingolipids with ceramide versus dihydroceramide backbones, and the ability to identify the ceramide backbone of sphingomyelins using a pseudo-MS3 protocol. Compounds that can be readily quantified using an internal standard cocktail developed by the LIPID MAPS Consortium are: sphingoid bases and sphingoid base 1-phosphates, more complex species such as ceramides, ceramide 1-phosphates, sphingomyelins, mono- and di-hexosylceramides, and these complex sphingolipids with dihydroceramide backbones. With minor modifications, glucosylceramides and galactosylceramides can be distinguished, and more complex species such as sulfatides can also be quantified, when the internal standards are available. LC ESI-MS/MS can be utilized to quantify a large number of structural and signaling sphingolipids using commercially available internal standards. The application of these methods is illustrated with RAW264.7 cells, a mouse macrophage cell line. These methods should be useful for a wide range of focused (sphingo)lipidomic investigations.
Figures








References
-
- Merrill A. H., Jr., Wang M. D., Park M., Sullards M. C. 2007. (Glyco)sphingolipidology: an amazing challenge and opportunity for systems biology. Trends Biochem. Sci. 32: 457–468 - PubMed
-
- van Echten-Deckert G.2000. Sphingolipid extraction and analysis by thin-layer chromatography. Methods Enzymol. 312: 64–79 - PubMed
-
- Muthing J.2000. Analyses of glycosphingolipids by high-performance liquid chromatography. Methods Enzymol. 312: 45–64 - PubMed
-
- Mano N., Oda Y., Yamada K., Asakawa N., Katayama K. 1997. Simultaneous quantitative determination method for sphingolipid metabolites by liquid chromatography/ionspray ionization tandem mass spectrometry. Anal. Biochem. 244: 291–300 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

