Eomesodermin requires transforming growth factor-beta/activin signaling and binds Smad2 to activate mesodermal genes
- PMID: 19036723
- PMCID: PMC2629102
- DOI: 10.1074/jbc.M808704200
Eomesodermin requires transforming growth factor-beta/activin signaling and binds Smad2 to activate mesodermal genes
Abstract
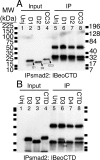
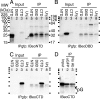
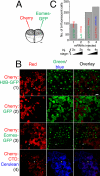
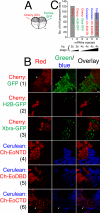
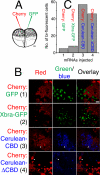
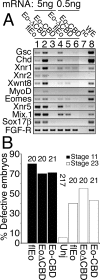
The T-box gene Eomesodermin (Eomes) is required for early embryonic mesoderm differentiation in mouse, frog (Xenopus laevis), and zebrafish, is important in late cardiac development in Xenopus, and for CD8+ T effector cell function in mouse. Eomes can ectopically activate many mesodermal genes. However, the mechanism by which Eomes activates transcription of these genes is poorly understood. We report that Eomes protein interacts with Smad2 and is capable of working in a non-cell autonomous manner via transfer of Eomes protein between adjacent embryonic cells. Blocking of Eomes protein transfer using a farnesylated red fluorescent protein (CherryF) also prevents Eomes nuclear accumulation. Transfer of Eomes protein between cells is mediated by the Eomes carboxyl terminus (456-692). A carbohydrate binding domain within the Eomes carboxyl-terminal region is sufficient for transfer and important for gene activation. We propose a novel mechanism by which Eomes helps effect a cellular response to a morphogen gradient.
Figures



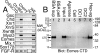
Similar articles
-
Tbx2 mediates dorsal patterning and germ layer suppression through inhibition of BMP/GDF and Activin/Nodal signaling.BMC Mol Cell Biol. 2020 May 28;21(1):39. doi: 10.1186/s12860-020-00282-1. BMC Mol Cell Biol. 2020. PMID: 32466750 Free PMC article.
-
A component of the ARC/Mediator complex required for TGF beta/Nodal signalling.Nature. 2002 Aug 8;418(6898):641-6. doi: 10.1038/nature00969. Epub 2002 Jul 24. Nature. 2002. PMID: 12167862
-
The Xenopus eomesodermin promoter and its concentration-dependent response to activin.Mech Dev. 2000 Jun;94(1-2):133-46. doi: 10.1016/s0925-4773(00)00300-2. Mech Dev. 2000. PMID: 10842065
-
Maternal control of pattern formation in Xenopus laevis.J Exp Zool B Mol Dev Evol. 2008 Jan 15;310(1):73-84. doi: 10.1002/jez.b.21153. J Exp Zool B Mol Dev Evol. 2008. PMID: 17219372 Review.
-
Forming and interpreting gradients in the early Xenopus embryo.Cold Spring Harb Perspect Biol. 2009 Jul;1(1):a002477. doi: 10.1101/cshperspect.a002477. Cold Spring Harb Perspect Biol. 2009. PMID: 20066079 Free PMC article. Review.
Cited by
-
Eomes function is conserved between zebrafish and mouse and controls left-right organiser progenitor gene expression via interlocking feedforward loops.Front Cell Dev Biol. 2022 Aug 25;10:982477. doi: 10.3389/fcell.2022.982477. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36133924 Free PMC article.
-
The T-box transcription factor Eomesodermin is essential for AVE induction in the mouse embryo.Genes Dev. 2013 May 1;27(9):997-1002. doi: 10.1101/gad.215152.113. Genes Dev. 2013. PMID: 23651855 Free PMC article.
-
Differential regulation of epiboly initiation and progression by zebrafish Eomesodermin A.Dev Biol. 2012 Feb 1;362(1):11-23. doi: 10.1016/j.ydbio.2011.10.036. Epub 2011 Nov 18. Dev Biol. 2012. PMID: 22142964 Free PMC article.
-
Pluripotency factors regulate definitive endoderm specification through eomesodermin.Genes Dev. 2011 Feb 1;25(3):238-50. doi: 10.1101/gad.607311. Epub 2011 Jan 18. Genes Dev. 2011. PMID: 21245162 Free PMC article.
-
Tbx2 mediates dorsal patterning and germ layer suppression through inhibition of BMP/GDF and Activin/Nodal signaling.BMC Mol Cell Biol. 2020 May 28;21(1):39. doi: 10.1186/s12860-020-00282-1. BMC Mol Cell Biol. 2020. PMID: 32466750 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

