Functional and structural characterization of the integrase from the prototype foamy virus
- PMID: 19036793
- PMCID: PMC2615609
- DOI: 10.1093/nar/gkn938
Functional and structural characterization of the integrase from the prototype foamy virus
Abstract
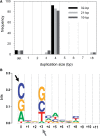
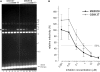
Establishment of the stable provirus is an essential step in retroviral replication, orchestrated by integrase (IN), a virus-derived enzyme. Until now, available structural information was limited to the INs of human immunodeficiency virus type 1 (HIV-1), avian sarcoma virus (ASV) and their close orthologs from the Lentivirus and Alpharetrovirus genera. Here, we characterized the in vitro activity of the prototype foamy virus (PFV) IN from the Spumavirus genus and determined the three-dimensional structure of its catalytic core domain (CCD). Recombinant PFV IN displayed robust and almost exclusively concerted integration activity in vitro utilizing donor DNA substrates as short as 16 bp, underscoring its significance as a model for detailed structural studies. Comparison of the HIV-1, ASV and PFV CCD structures highlighted both conserved as well as unique structural features such as organization of the active site and the putative host factor binding face. Despite possessing very limited sequence identity to its HIV counterpart, PFV IN was sensitive to HIV IN strand transfer inhibitors, suggesting that this class of inhibitors target the most conserved features of retroviral IN-DNA complexes.
Figures



References
-
- Asante-Appiah E, Skalka AM. HIV-1 integrase: structural organization, conformational changes, and catalysis. Adv. Virus Res. 1999;52:351–369. - PubMed
-
- Craigie R. Retroviral DNA integration. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington DC: ASM Press; 2002. pp. 613–630.
-
- Lewinski MK, Bushman FD. Retroviral DNA integration—mechanism and consequences. Adv. Genet. 2005;55:147–181. - PubMed
-
- Enssle J, Moebes A, Heinkelein M, Panhuysen M, Mauer B, Schweizer M, Neumann-Haefelin D, Rethwilm A. An active foamy virus integrase is required for virus replication. J. Gen. Virol. 1999;80:1445–1452. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

