Distinct domains within APOBEC3G and APOBEC3F interact with separate regions of human immunodeficiency virus type 1 Vif
- PMID: 19036809
- PMCID: PMC2643761
- DOI: 10.1128/JVI.01621-08
Distinct domains within APOBEC3G and APOBEC3F interact with separate regions of human immunodeficiency virus type 1 Vif
Abstract
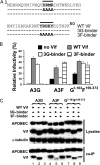
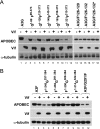
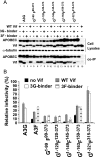
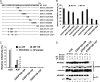
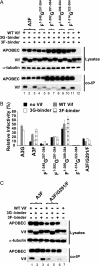
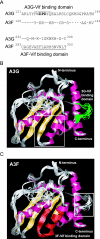
Human APOBEC3G (A3G) and APOBEC3F (A3F) inhibit the replication of Vif-deficient human immunodeficiency virus type 1 (HIV-1). HIV-1 Vif overcomes these host restriction factors by binding to them and inducing their degradation. Thus, the Vif-A3G and Vif-A3F interactions are attractive targets for antiviral drug development, as inhibiting these interactions could allow the host defense mechanism to control HIV-1 replication. Recently, it has been reported that amino acids 105 to 156 of A3G are involved in the interaction with Vif; however, to date, the region of A3F involved in Vif binding has not been identified. Using our previously reported Vif mutants that are capable of binding to only A3G (3G binder) or only A3F (3F binder), in conjunction with a series of A3G-A3F chimeras, we have now mapped the APOBEC3-Vif interaction domains. We found that the A3G domain that interacts with the Vif YRHHY region is located between amino acids 126 and 132 of A3G, which is consistent with the conclusions reported in previous studies. The A3F domain that interacts with the Vif DRMR region did not occur in the homologous domain but instead was located between amino acids 283 and 300 of A3F. These studies are the first to identify the A3F domain that interacts with the Vif DRMR region and show that distinct domains of A3G and A3F interact with different Vif regions. Pharmacological inhibition of either or both of these Vif-A3 interactions should prevent the degradation of the APOBEC3 proteins and could be used as a therapy against HIV-1.
Figures

Similar articles
-
Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F.J Virol. 2007 Aug;81(15):8201-10. doi: 10.1128/JVI.00395-07. Epub 2007 May 23. J Virol. 2007. PMID: 17522216 Free PMC article.
-
Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction.J Mol Biol. 2008 Sep 12;381(4):1000-11. doi: 10.1016/j.jmb.2008.06.061. Epub 2008 Jun 28. J Mol Biol. 2008. PMID: 18619467
-
Mechanism of Enhanced HIV Restriction by Virion Coencapsidated Cytidine Deaminases APOBEC3F and APOBEC3G.J Virol. 2017 Jan 18;91(3):e02230-16. doi: 10.1128/JVI.02230-16. Print 2017 Feb 1. J Virol. 2017. PMID: 27881650 Free PMC article.
-
Multiple ways of targeting APOBEC3-virion infectivity factor interactions for anti-HIV-1 drug development.Trends Pharmacol Sci. 2009 Dec;30(12):638-46. doi: 10.1016/j.tips.2009.09.006. Trends Pharmacol Sci. 2009. PMID: 19837465 Free PMC article. Review.
-
Various strategies for developing APOBEC3G protectors to circumvent human immunodeficiency virus type 1.Eur J Med Chem. 2023 Mar 15;250:115188. doi: 10.1016/j.ejmech.2023.115188. Epub 2023 Feb 6. Eur J Med Chem. 2023. PMID: 36773550 Review.
Cited by
-
A role for gorilla APOBEC3G in shaping lentivirus evolution including transmission to humans.PLoS Pathog. 2020 Sep 10;16(9):e1008812. doi: 10.1371/journal.ppat.1008812. eCollection 2020 Sep. PLoS Pathog. 2020. PMID: 32913367 Free PMC article.
-
Small molecular compounds inhibit HIV-1 replication through specifically stabilizing APOBEC3G.J Biol Chem. 2010 May 28;285(22):16546-52. doi: 10.1074/jbc.M109.085308. Epub 2010 Apr 2. J Biol Chem. 2010. PMID: 20363737 Free PMC article.
-
Signals in APOBEC3F N-terminal and C-terminal deaminase domains each contribute to encapsidation in HIV-1 virions and are both required for HIV-1 restriction.J Biol Chem. 2012 May 11;287(20):16965-74. doi: 10.1074/jbc.M111.310839. Epub 2012 Mar 28. J Biol Chem. 2012. PMID: 22451677 Free PMC article.
-
A patch of positively charged amino acids surrounding the human immunodeficiency virus type 1 Vif SLVx4Yx9Y motif influences its interaction with APOBEC3G.J Virol. 2009 Sep;83(17):8674-82. doi: 10.1128/JVI.00653-09. Epub 2009 Jun 17. J Virol. 2009. PMID: 19535450 Free PMC article.
-
APOBEC3 degradation is the primary function of HIV-1 Vif for virus replication in the myeloid cell line THP-1.bioRxiv [Preprint]. 2023 Mar 29:2023.03.28.534666. doi: 10.1101/2023.03.28.534666. bioRxiv. 2023. Update in: mBio. 2023 Aug 31;14(4):e0078223. doi: 10.1128/mbio.00782-23. PMID: 37034786 Free PMC article. Updated. Preprint.
References
-
- Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 141392-1396. - PubMed
-
- Chen, K. M., E. Harjes, P. J. Gross, A. Fahmy, Y. Lu, K. Shindo, R. S. Harris, and H. Matsuo. 2008. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature 452116-119. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

