Pil1 controls eisosome biogenesis
- PMID: 19037108
- PMCID: PMC2633383
- DOI: 10.1091/mbc.e08-03-0313
Pil1 controls eisosome biogenesis
Abstract
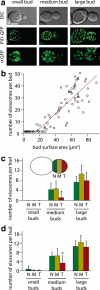
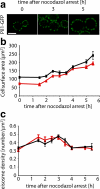
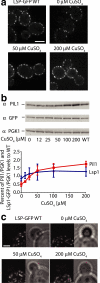
The molecular composition of plasma membranes is constantly remodeled by endocytosis and exocytosis. Eisosomes are large cytoplasmic protein assemblies that localize to specialized domains on the yeast plasma membrane. They are of uniform size and immobile, and their disruption leads to large aberrant plasma membrane invaginations and endocytic defects. It is unknown how eisosomes are formed or inherited and what governs their size, distribution, and location. Here we show that eisosomes are formed de novo in the bud of dividing cells. They colonize newly formed membrane at a fixed density in a polarized wave proceeding from the bud neck to the bud tip and become anchored at the site of their formation. Pil1, one of the two main eisosome subunits, emerges as the central regulator of eisosome biogenesis that determines both size and location of eisosomes. Lowering Pil1 expression leads to normal-sized eisosomes at a reduced density, suggesting that eisosomes must be of a minimal size. Conversely, raising Pil1 expression leads to larger eisosomes at a fixed density, suggesting that under these conditions eisosome nucleation sites are limiting. Pil1 expression is regulated by the cell cycle, which synchronizes eisosome formation with plasma membrane growth. Our results establish a first framework of the molecular principles that define eisosome assembly and distribution.
Figures






Similar articles
-
Eisosomes are dynamic plasma membrane domains showing pil1-lsp1 heteroligomer binding equilibrium.Biophys J. 2015 Apr 7;108(7):1633-1644. doi: 10.1016/j.bpj.2015.02.011. Biophys J. 2015. PMID: 25863055 Free PMC article.
-
Lsp1 partially substitutes for Pil1 function in eisosome assembly under stress conditions.J Cell Sci. 2023 Feb 1;136(3):jcs260554. doi: 10.1242/jcs.260554. Epub 2023 Feb 6. J Cell Sci. 2023. PMID: 36601791
-
Seg1 controls eisosome assembly and shape.J Cell Biol. 2012 Aug 6;198(3):405-20. doi: 10.1083/jcb.201202097. J Cell Biol. 2012. PMID: 22869600 Free PMC article.
-
Insights into eisosome assembly and organization.J Biosci. 2012 Jun;37(2):295-500. doi: 10.1007/s12038-012-9206-6. J Biosci. 2012. PMID: 22581335 Review.
-
Plasma Membrane MCC/Eisosome Domains Promote Stress Resistance in Fungi.Microbiol Mol Biol Rev. 2020 Sep 16;84(4):e00063-19. doi: 10.1128/MMBR.00063-19. Print 2020 Nov 18. Microbiol Mol Biol Rev. 2020. PMID: 32938742 Free PMC article. Review.
Cited by
-
Effects of FSGS-associated mutations on the stability and function of myosin-1 in fission yeast.Dis Model Mech. 2015 Aug 1;8(8):891-902. doi: 10.1242/dmm.020214. Epub 2015 Jun 18. Dis Model Mech. 2015. PMID: 26092123 Free PMC article.
-
Tetraspanner-based nanodomains modulate BAR domain-induced membrane curvature.EMBO Rep. 2023 Dec 6;24(12):e57232. doi: 10.15252/embr.202357232. Epub 2023 Oct 30. EMBO Rep. 2023. PMID: 37902009 Free PMC article.
-
The joint action of yeast eisosomes and membraneless organelles in response to ethanol stress.Heliyon. 2024 May 19;10(10):e31561. doi: 10.1016/j.heliyon.2024.e31561. eCollection 2024 May 30. Heliyon. 2024. PMID: 38818138 Free PMC article.
-
Microdomain Protein Nce102 Is a Local Sensor of Plasma Membrane Sphingolipid Balance.Microbiol Spectr. 2022 Aug 31;10(4):e0196122. doi: 10.1128/spectrum.01961-22. Epub 2022 Jun 27. Microbiol Spectr. 2022. PMID: 35758748 Free PMC article.
-
TORC2 and eisosomes are spatially interdependent, requiring optimal level of phosphatidylinositol 4, 5-bisphosphate for their integrity.J Biosci. 2015 Jun;40(2):299-311. doi: 10.1007/s12038-015-9526-4. J Biosci. 2015. PMID: 25963258
References
-
- Bacon D. W., Watts D. G. Estimating the transition between two intersecting straight lines. Biometrika. 1971;58:525–534.
-
- Bishop A. C., et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. - PubMed
-
- Broadie K. Synapse scaffolding: intersection of endocytosis and growth. Curr. Biol. 2004;14:R853–R855. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

