Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology
- PMID: 19037247
- PMCID: PMC2742159
- DOI: 10.1038/nature07541
Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology
Abstract
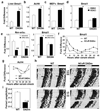
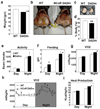
Rhythmic changes in histone acetylation at circadian clock genes suggest that temporal modulation of gene expression is regulated by chromatin modifications. Furthermore, recent studies demonstrate a critical relationship between circadian and metabolic physiology. The nuclear receptor corepressor 1 (Ncor1) functions as an activating subunit for the chromatin modifying enzyme histone deacetylase 3 (Hdac3). Lack of Ncor1 is incompatible with life, and hence it is unknown whether Ncor1, and particularly its regulation of Hdac3, is critical for adult mammalian physiology. Here we show that specific, genetic disruption of the Ncor1-Hdac3 interaction in mice causes aberrant regulation of clock genes and results in abnormal circadian behaviour. These mice are also leaner and more insulin-sensitive owing to increased energy expenditure. Unexpectedly, loss of a functional Ncor1-Hdac3 complex in vivo does not lead to sustained increases in known catabolic genes, but instead significantly alters the oscillatory patterns of several metabolic genes, demonstrating that circadian regulation of metabolism is critical for normal energy balance. These findings indicate that activation of Hdac3 by Ncor1 is a nodal point in the epigenetic regulation of circadian and metabolic physiology.
Figures


Comment in
-
Circadian clocks: tips from the tip of the iceberg.Nature. 2008 Dec 18;456(7224):881-3. doi: 10.1038/456881a. Nature. 2008. PMID: 19092918 No abstract available.
References
-
- Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–374. - PubMed
-
- Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. - PubMed
-
- Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. - PubMed
-
- Kohsaka A, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

