VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse
- PMID: 19037259
- PMCID: PMC2600653
- DOI: 10.1038/emboj.2008.248
VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse
Abstract
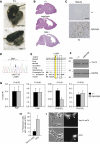
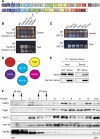
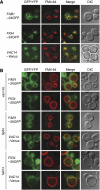
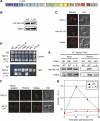
The signalling lipid PI(3,5)P(2) is generated on endosomes and regulates retrograde traffic to the trans-Golgi network. Physiological signals regulate rapid, transient changes in PI(3,5)P(2) levels. Mutations that lower PI(3,5)P(2) cause neurodegeneration in human patients and mice. The function of Vac14 in the regulation of PI(3,5)P(2) was uncharacterized previously. Here, we predict that yeast and mammalian Vac14 are composed entirely of HEAT repeats and demonstrate that Vac14 exerts an effect as a scaffold for the PI(3,5)P(2) regulatory complex by direct contact with the known regulators of PI(3,5)P(2): Fig4, Fab1, Vac7 and Atg18. We also report that the mouse mutant ingls (infantile gliosis) results from a missense mutation in Vac14 that prevents the association of Vac14 with Fab1, generating a partial complex. Analysis of ingls and two additional mutants provides insight into the organization of the PI(3,5)P(2) regulatory complex and indicates that Vac14 mediates three distinct mechanisms for the rapid interconversion of PI3P and PI(3,5)P(2). Moreover, these studies show that the association of Fab1 with the complex is essential for viability in the mouse.
Figures
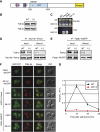
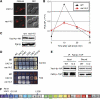

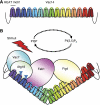
Comment in
-
A protein complex that regulates PtdIns(3,5)P2 levels.EMBO J. 2009 Jan 21;28(2):86-7. doi: 10.1038/emboj.2008.270. EMBO J. 2009. PMID: 19158662 Free PMC article.
References
-
- Andrade MA, Ponting CP, Gibson TJ, Bork P (2000) Homology-based method for identification of protein repeats using statistical significance estimates. J Mol Biol 298: 521–537 - PubMed
-
- Behnia R, Munro S (2005) Organelle identity and the signposts for membrane traffic. Nature 438: 597–604 - PubMed
-
- Bolis A, Zordan P, Coviello S, Bolino A (2007) Myotubularin-related (MTMR) phospholipid phosphatase proteins in the peripheral nervous system. Mol Neurobiol 35: 308–316 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

