The Protein Model Portal
- PMID: 19037750
- PMCID: PMC2704613
- DOI: 10.1007/s10969-008-9048-5
The Protein Model Portal
Abstract
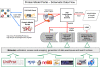
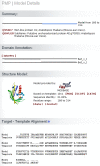
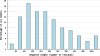
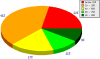
Structural Genomics has been successful in determining the structures of many unique proteins in a high throughput manner. Still, the number of known protein sequences is much larger than the number of experimentally solved protein structures. Homology (or comparative) modeling methods make use of experimental protein structures to build models for evolutionary related proteins. Thereby, experimental structure determination efforts and homology modeling complement each other in the exploration of the protein structure space. One of the challenges in using model information effectively has been to access all models available for a specific protein in heterogeneous formats at different sites using various incompatible accession code systems. Often, structure models for hundreds of proteins can be derived from a given experimentally determined structure, using a variety of established methods. This has been done by all of the PSI centers, and by various independent modeling groups. The goal of the Protein Model Portal (PMP) is to provide a single portal which gives access to the various models that can be leveraged from PSI targets and other experimental protein structures. A single interface allows all existing pre-computed models across these various sites to be queried simultaneously, and provides links to interactive services for template selection, target-template alignment, model building, and quality assessment. The current release of the portal consists of 7.6 million model structures provided by different partner resources (CSMP, JCSG, MCSG, NESG, NYSGXRC, JCMM, ModBase, SWISS-MODEL Repository). The PMP is available at http://www.proteinmodelportal.org and from the PSI Structural Genomics Knowledgebase.
Figures


References
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1093/nar/gkl971', 'is_inner': False, 'url': 'https://doi.org/10.1093/nar/gkl971'}, {'type': 'PMC', 'value': 'PMC1669775', 'is_inner': False, 'url': 'https://pmc.ncbi.nlm.nih.gov/articles/PMC1669775/'}, {'type': 'PubMed', 'value': '17142228', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/17142228/'}]}
- Berman H et al (2007) The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res 35:D301–D303. doi:10.1093/nar/gkl971 - PMC - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1371/journal.pbio.0050016', 'is_inner': False, 'url': 'https://doi.org/10.1371/journal.pbio.0050016'}, {'type': 'PMC', 'value': 'PMC1821046', 'is_inner': False, 'url': 'https://pmc.ncbi.nlm.nih.gov/articles/PMC1821046/'}, {'type': 'PubMed', 'value': '17355171', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/17355171/'}]}
- Yooseph S et al (2007) The sorcerer II global ocean sampling expedition: expanding the universe of protein families. PLoS Biol 5:e16. doi:10.1371/journal.pbio.0050016 - PMC - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1126/science.1065659', 'is_inner': False, 'url': 'https://doi.org/10.1126/science.1065659'}, {'type': 'PubMed', 'value': '11588250', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/11588250/'}]}
- Baker D, Sali A (2001) Protein structure prediction and structural genomics. Science 294:93–96. doi:10.1126/science.1065659 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1016/S1359-6446(04)03196-4', 'is_inner': False, 'url': 'https://doi.org/10.1016/s1359-6446(04)03196-4'}, {'type': 'PMC', 'value': 'PMC7129151', 'is_inner': False, 'url': 'https://pmc.ncbi.nlm.nih.gov/articles/PMC7129151/'}, {'type': 'PubMed', 'value': '15279849', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/15279849/'}]}
- Hillisch A, Pineda LF, Hilgenfeld R (2004) Utility of homology models in the drug discovery process. Drug Discov Today 9:659–669. doi:10.1016/S1359-6446(04)03196-4 - PMC - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1093/bioinformatics/18.7.934', 'is_inner': False, 'url': 'https://doi.org/10.1093/bioinformatics/18.7.934'}, {'type': 'PubMed', 'value': '12117790', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/12117790/'}]}
- Peitsch MC (2002) About the use of protein models. Bioinformatics 18:934–938. doi:10.1093/bioinformatics/18.7.934 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

