PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit
- PMID: 19038223
- PMCID: PMC2734413
- DOI: 10.1016/j.neuron.2008.10.042
PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit
Erratum in
- Neuron. 2009 Jan 15;61(1):152. Kang, Keongjin [corrected to Kang, Kyeongjin]
Abstract
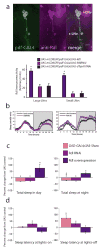
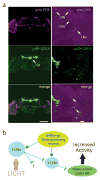
Daily sleep cycles in humans are driven by a complex circuit within which GABAergic sleep-promoting neurons oppose arousal. Drosophila sleep has recently been shown to be controlled by GABA, which acts on unknown cells expressing the Rdl GABAA receptor. We identify here the relevant Rdl-containing cells as PDF-expressing small and large ventral lateral neurons (LNvs) of the circadian clock. LNv activity regulates total sleep as well as the rate of sleep onset; both large and small LNvs are part of the sleep circuit. Flies mutant for pdf or its receptor are hypersomnolent, and PDF acts on the LNvs themselves to control sleep. These features of the Drosophila sleep circuit, GABAergic control of onset and maintenance as well as peptidergic control of arousal, support the idea that features of sleep-circuit architecture as well as the mechanisms governing the behavioral transitions between sleep and wake are conserved between mammals and insects.
Figures




References
-
- Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. - PubMed
-
- Borbely AA, Achermann P. Sleep homeostasis and models of sleep regulation. J Biol Rhythms. 1999;14:557–568. - PubMed
-
- Broughton SJ, Kitamoto T, Greenspan RJ. Excitatory and inhibitory switches for courtship in the brain of Drosophila melanogaster. Curr Biol. 2004;14:538–547. - PubMed
-
- Choi JC, Park D, Griffith LC. Electrophysiological and morphological characterization of identified motor neurons in the Drosophila third instar larva central nervous system. J Neurophysiol. 2004;91:2353–2365. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

