Ligand-induced ErbB receptor dimerization
- PMID: 19038249
- PMCID: PMC2667204
- DOI: 10.1016/j.yexcr.2008.10.024
Ligand-induced ErbB receptor dimerization
Abstract
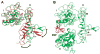
Structural studies have provided important new insights into how ligand binding promotes homodimerization and activation of the EGF receptor and the other members of the ErbB family of receptor tyrosine kinases. These structures have also suggested possible explanations for the unique properties of ErbB2, which has no known ligand and can cause cell transformation (and tumorigenesis) by simple overexpression. In parallel with these advances, studies of the EGF receptor at the cell surface increasingly argue that the structural studies are missing key mechanistic components. This is particularly evident in the structural prediction that EGF binding linked to receptor dimerization should be positively cooperative, whereas cell-surface EGF-binding studies suggest negative cooperativity. In this review, I summarize studies of ErbB receptor extracellular regions in solution and of intact receptors at the cell surface, and attempt to reconcile the differences suggested by the two approaches. By combining results obtained with receptor 'parts', it is qualitatively possible to explain some models for the properties of the whole receptor. These considerations underline the need to consider the intact ErbB receptors as intact allosterically regulated enzymes, and to combine cellular and structural studies into a complete picture.
Figures

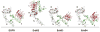

References
-
- Ullrich A, Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell. 1990;61:203–212. - PubMed
-
- Yarden Y, Schlessinger J. Epidermal growth factor induces rapid, reversible aggregation of the purified epidermal growth factor receptor. Biochemistry. 1987;26:1443–1451. - PubMed
-
- Yarden Y, Schlessinger J. Self-phosphorylation of epidermal growth factor receptor: evidence for a model of intermolecular allosteric activation. Biochemistry. 1987;26:1434–1442. - PubMed
-
- Jorissen RN, Walker F, Pouliot N, Garrett TPJ, Ward CW, Burgess AW. Epidermal growth factor receptor: mechanisms of activation and signalling. Exp Cell Res. 2003;284:31–53. - PubMed
-
- Holbro T, Hynes NE. ErbB receptors: directing key signaling networks throughout life. Annu Rev Pharmacol Toxicol. 2004;44:195–217. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

