Automated mapping of hippocampal atrophy in 1-year repeat MRI data from 490 subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls
- PMID: 19041724
- PMCID: PMC2733354
- DOI: 10.1016/j.neuroimage.2008.10.043
Automated mapping of hippocampal atrophy in 1-year repeat MRI data from 490 subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls
Abstract
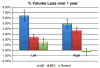
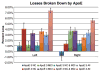
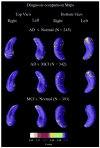
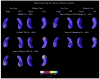
As one of the earliest structures to degenerate in Alzheimer's disease (AD), the hippocampus is the target of many studies of factors that influence rates of brain degeneration in the elderly. In one of the largest brain mapping studies to date, we mapped the 3D profile of hippocampal degeneration over time in 490 subjects scanned twice with brain MRI over a 1-year interval (980 scans). We examined baseline and 1-year follow-up scans of 97 AD subjects (49 males/48 females), 148 healthy control subjects (75 males/73 females), and 245 subjects with mild cognitive impairment (MCI; 160 males/85 females). We used our previously validated automated segmentation method, based on AdaBoost, to create 3D hippocampal surface models in all 980 scans. Hippocampal volume loss rates increased with worsening diagnosis (normal=0.66%/year; MCI=3.12%/year; AD=5.59%/year), and correlated with both baseline and interval changes in Mini-Mental State Examination (MMSE) scores and global and sum-of-boxes Clinical Dementia Rating scale (CDR) scores. Surface-based statistical maps visualized a selective profile of ongoing atrophy in all three diagnostic groups. Healthy controls carrying the ApoE4 gene atrophied faster than non-carriers, while more educated controls atrophied more slowly; converters from MCI to AD showed faster atrophy than non-converters. Hippocampal loss rates can be rapidly mapped, and they track cognitive decline closely enough to be used as surrogate markers of Alzheimer's disease in drug trials. They also reveal genetically greater atrophy in cognitively intact subjects.
Conflict of interest statement
Conflict of interest
The authors declare that there are no conflicts of interest.
Figures




References
-
- Alexander GE, Chen K, Merkley TL, Reiman EM, Caselli RJ, Aschenbrenner M, Santerre-Lemmon L, Lewis DJ, Pietrini P, Teipel SJ, Hampel H, Rapoport SI, Moeller JR. Regional network of magnetic resonance imaging gray matter volume in healthy aging. NeuroReport. 2006;17:951–956. - PubMed
-
- Apostolova LG, Dinov ID, Dutton RA, Hayashi KM, Toga AW, Cummings JL, Thompson PM. 3D comparison of hippocampal atrophy in amnestic mild cognitive impairment and Alzheimer’s disease. Brain. 2006a;129:2867–2873. - PubMed
-
- Apostolova LG, Dutton RA, Dinov ID, Hayashi KM, Toga AW, Cummings JL, Thompson PM. Conversion of mild cognitive impairment to Alzheimer disease predicted by hippocampal atrophy maps. Arch Neurol. 2006b;63:693–699. - PubMed
-
- Apostolova LG, Akopyan GG, Partiali N, Steiner CA, Dutton RA, Hayashi KM, Dinov ID, Toga AW, Cummings JL, Thompson PM. Structural correlates of apathy in Alzheimer’s disease. Dement Geriatr Cogn Disord. 2007;24:91–97. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

