Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance
- PMID: 19041766
- PMCID: PMC2652870
- DOI: 10.1016/j.cmet.2008.10.008
Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance
Abstract
Animal cells control their membrane lipid composition within narrow limits, but the sensing mechanisms underlying this control are largely unknown. Recent studies disclosed a protein network that controls the level of one lipid-cholesterol. This network resides in the endoplasmic reticulum (ER). A key component is Scap, a tetrameric ER membrane protein that binds cholesterol. Cholesterol binding prevents Scap from transporting SREBPs to the Golgi for activation. Using a new method to purify ER membranes from cultured cells, we show that Scap responds cooperatively to ER cholesterol levels. When ER cholesterol exceeds 5% of total ER lipids (molar basis), SREBP-2 transport is abruptly blocked. Transport resumes when ER cholesterol falls below the 5% threshold. The 5% threshold is lowered to 3% when cells overexpress Insig-1, a Scap-binding protein. Cooperative interactions between cholesterol, Scap, and Insig create a sensitive switch that controls the cholesterol composition of cell membranes with remarkable precision.
Figures

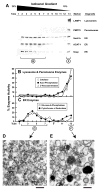

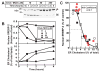


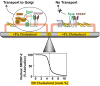
Comment in
-
A cholesterol toggle switch.Cell Metab. 2008 Dec;8(6):451-3. doi: 10.1016/j.cmet.2008.11.006. Cell Metab. 2008. PMID: 19041760
References
-
- Adams CM, Reitz J, DeBrabander JK, Feramisco JD, Brown MS, Goldstein JL. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J Biol Chem. 2004;279:52772–52780. - PubMed
-
- Balch WE, Rothman JE. Characterization of protein transport between successive compartments of the Golgi apparatus: Asymmetric properties of donor and acceptor activities in a cell-free system. Arch Biochem Biophysics. 1985;240:413–425. - PubMed
-
- Bergstrand A, Dallner G. Isolation of rough and smooth microsomes from rat liver by means of a commercially available centrifuge. Analytical Biochem. 1969;29:351–356. - PubMed
-
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. - PubMed
-
- Brown AJ, Sun L, Feramisco JD, Brown MS, Goldstein JL. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol Cell. 2002;10:237–245. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

