Rex1p deficiency leads to accumulation of precursor initiator tRNAMet and polyadenylation of substrate RNAs in Saccharomyces cerevisiae
- PMID: 19042972
- PMCID: PMC2615624
- DOI: 10.1093/nar/gkn925
Rex1p deficiency leads to accumulation of precursor initiator tRNAMet and polyadenylation of substrate RNAs in Saccharomyces cerevisiae
Abstract
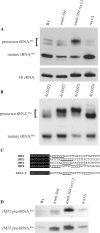
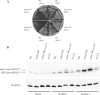
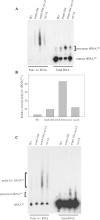
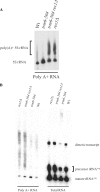
A synthetic genetic array was used to identify lethal and slow-growth phenotypes produced when a mutation in TRM6, which encodes a tRNA modification enzyme subunit, was combined with the deletion of any non-essential gene in Saccharomyces cerevisiae. We found that deletion of the REX1 gene resulted in a slow-growth phenotype in the trm6-504 strain. Previously, REX1 was shown to be involved in processing the 3' ends of 5S rRNA and the dimeric tRNA(Arg)-tRNA(Asp). In this study, we have discovered a requirement for Rex1p in processing the 3' end of tRNA(i)(Met) precursors and show that precursor tRNA(i)(Met) accumulates in a trm6-504 rex1Delta strain. Loss of Rex1p results in polyadenylation of its substrates, including tRNA(i)(Met), suggesting that defects in 3' end processing can activate the nuclear surveillance pathway. Finally, purified Rex1p displays Mg(2+)-dependent ribonuclease activity in vitro, and the enzyme is inactivated by mutation of two highly conserved amino acids.
Figures

Similar articles
-
Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae.Genes Dev. 2004 Jun 1;18(11):1227-40. doi: 10.1101/gad.1183804. Epub 2004 May 14. Genes Dev. 2004. PMID: 15145828 Free PMC article.
-
Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA.RNA. 2006 Mar;12(3):508-21. doi: 10.1261/rna.2305406. Epub 2006 Jan 23. RNA. 2006. PMID: 16431988 Free PMC article.
-
Competition between the Rex1 exonuclease and the La protein affects both Trf4p-mediated RNA quality control and pre-tRNA maturation.RNA. 2008 Jun;14(6):1214-27. doi: 10.1261/rna.1050408. Epub 2008 May 2. RNA. 2008. PMID: 18456844 Free PMC article.
-
3' processing of eukaryotic precursor tRNAs.Wiley Interdiscip Rev RNA. 2011 May-Jun;2(3):362-75. doi: 10.1002/wrna.64. Wiley Interdiscip Rev RNA. 2011. PMID: 21572561 Free PMC article. Review.
-
Two-subunit enzymes involved in eukaryotic post-transcriptional tRNA modification.RNA Biol. 2014;11(12):1608-18. doi: 10.1080/15476286.2015.1008360. RNA Biol. 2014. PMID: 25625329 Free PMC article. Review.
Cited by
-
Maf1-mediated regulation of yeast RNA polymerase III is correlated with CCA addition at the 3' end of tRNA precursors.Gene. 2017 May 15;612:12-18. doi: 10.1016/j.gene.2016.08.033. Epub 2016 Aug 27. Gene. 2017. PMID: 27575455 Free PMC article.
-
Methodology for the High-Throughput Identification and Characterization of tRNA Variants That Are Substrates for a tRNA Decay Pathway.Methods Enzymol. 2015;560:1-17. doi: 10.1016/bs.mie.2015.03.003. Epub 2015 Apr 27. Methods Enzymol. 2015. PMID: 26253963 Free PMC article.
-
Contribution of domain structure to the function of the yeast DEDD family exoribonuclease and RNase T functional homolog, Rex1.RNA. 2022 Apr;28(4):493-507. doi: 10.1261/rna.078939.121. Epub 2022 Jan 26. RNA. 2022. PMID: 35082142 Free PMC article.
-
Cellular dynamics of tRNAs and their genes.FEBS Lett. 2010 Jan 21;584(2):310-7. doi: 10.1016/j.febslet.2009.11.053. FEBS Lett. 2010. PMID: 19931532 Free PMC article. Review.
-
Conserved and divergent features of the structure and function of La and La-related proteins (LARPs).Biochim Biophys Acta. 2010 May-Jun;1799(5-6):365-78. doi: 10.1016/j.bbagrm.2010.01.011. Epub 2010 Feb 2. Biochim Biophys Acta. 2010. PMID: 20138158 Free PMC article. Review.
References
-
- Wolin SL, Matera AG. The trials and travels of tRNA. Genes Dev. 1999;13:1–10. - PubMed
-
- Björk GR. In: tRNA: Structure Biosynthesis and Function. Söll D, RajBhandary UL, editors. Washington, D.C: ASM Press; 1995. pp. 165–206.
-
- Schürer H, Schiffer S, Marchfelder A, Mörl M. This Is the End: Processing, Editing, and Repair at the tRNA 3′-Terminus. Biol. Chem. 2001;382:1147–1156. - PubMed
-
- Li Z, Deutscher MP. Maturation pathways for E. coli tRNA precursors: a random multienzyme process in vivo. Cell. 1996;86:503–512. - PubMed
-
- Papadimitriou A, Gross HJ. Pre-tRNA 3′-processing in Saccharomyces cerevisiae. Purification and characterization of exo- and endoribonucleases. Eur. J. Biochem. 1996;242:747–759. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

