Profiling synaptic proteins identifies regulators of insulin secretion and lifespan
- PMID: 19043554
- PMCID: PMC2582949
- DOI: 10.1371/journal.pgen.1000283
Profiling synaptic proteins identifies regulators of insulin secretion and lifespan
Abstract
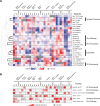
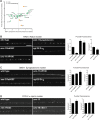
Cells are organized into distinct compartments to perform specific tasks with spatial precision. In neurons, presynaptic specializations are biochemically complex subcellular structures dedicated to neurotransmitter secretion. Activity-dependent changes in the abundance of presynaptic proteins are thought to endow synapses with different functional states; however, relatively little is known about the rules that govern changes in the composition of presynaptic terminals. We describe a genetic strategy to systematically analyze protein localization at Caenorhabditis elegans presynaptic specializations. Nine presynaptic proteins were GFP-tagged, allowing visualization of multiple presynaptic structures. Changes in the distribution and abundance of these proteins were quantified in 25 mutants that alter different aspects of neurotransmission. Global analysis of these data identified novel relationships between particular presynaptic components and provides a new method to compare gene functions by identifying shared protein localization phenotypes. Using this strategy, we identified several genes that regulate secretion of insulin-like growth factors (IGFs) and influence lifespan in a manner dependent on insulin/IGF signaling.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

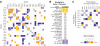



References
-
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. - PubMed
-
- Piano F, Schetter AJ, Morton DG, Gunsalus KC, Reinke V, et al. Gene clustering based on RNAi phenotypes of ovary-enriched genes in C. elegans. Curr Biol. 2002;12:1959–1964. - PubMed
-
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. - PubMed
-
- Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–547. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

