Acquisition and evolution of plant pathogenesis-associated gene clusters and candidate determinants of tissue-specificity in xanthomonas
- PMID: 19043590
- PMCID: PMC2585010
- DOI: 10.1371/journal.pone.0003828
Acquisition and evolution of plant pathogenesis-associated gene clusters and candidate determinants of tissue-specificity in xanthomonas
Abstract
Background: Xanthomonas is a large genus of plant-associated and plant-pathogenic bacteria. Collectively, members cause diseases on over 392 plant species. Individually, they exhibit marked host- and tissue-specificity. The determinants of this specificity are unknown.
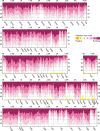
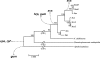
Methodology/principal findings: To assess potential contributions to host- and tissue-specificity, pathogenesis-associated gene clusters were compared across genomes of eight Xanthomonas strains representing vascular or non-vascular pathogens of rice, brassicas, pepper and tomato, and citrus. The gum cluster for extracellular polysaccharide is conserved except for gumN and sequences downstream. The xcs and xps clusters for type II secretion are conserved, except in the rice pathogens, in which xcs is missing. In the otherwise conserved hrp cluster, sequences flanking the core genes for type III secretion vary with respect to insertion sequence element and putative effector gene content. Variation at the rpf (regulation of pathogenicity factors) cluster is more pronounced, though genes with established functional relevance are conserved. A cluster for synthesis of lipopolysaccharide varies highly, suggesting multiple horizontal gene transfers and reassortments, but this variation does not correlate with host- or tissue-specificity. Phylogenetic trees based on amino acid alignments of gum, xps, xcs, hrp, and rpf cluster products generally reflect strain phylogeny. However, amino acid residues at four positions correlate with tissue specificity, revealing hpaA and xpsD as candidate determinants. Examination of genome sequences of xanthomonads Xylella fastidiosa and Stenotrophomonas maltophilia revealed that the hrp, gum, and xcs clusters are recent acquisitions in the Xanthomonas lineage.
Conclusions/significance: Our results provide insight into the ancestral Xanthomonas genome and indicate that differentiation with respect to host- and tissue-specificity involved not major modifications or wholesale exchange of clusters, but subtle changes in a small number of genes or in non-coding sequences, and/or differences outside the clusters, potentially among regulatory targets or secretory substrates.
Conflict of interest statement
Figures


References
-
- Perna NT, Plunkett G, III, Burland V, Mau B, Glasner JD, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. - PubMed
-
- Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. - PubMed
-
- Brosch R, Pym AS, Gordon SV, Cole ST. The evolution of mycobacterial pathogenicity: clues from comparative genomics. Trends Microbiol. 2001;9:452–458. - PubMed
-
- Raskin DM, Seshadri R, Pukatzki SU, Mekalanos JJ. Bacterial genomics and pathogen evolution. Cell. 2006;124:703–714. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

