Perilobar nephrogenic rests are nonobligate molecular genetic precursor lesions of insulin-like growth factor-II-associated Wilms tumors
- PMID: 19047088
- PMCID: PMC2659869
- DOI: 10.1158/1078-0432.CCR-08-1620
Perilobar nephrogenic rests are nonobligate molecular genetic precursor lesions of insulin-like growth factor-II-associated Wilms tumors
Abstract
Purpose: Perilobar nephrogenic rests (PLNRs) are abnormally persistent foci of embryonal immature blastema that have been associated with dysregulation at the 11p15 locus by genetic/epigenetic means and are thought to be precursor lesions of Wilms tumor. The precise genomic events are, however, largely unknown.
Experimental design: We used array comparative genomic hybridization to analyze a series of 50 PLNRs and 25 corresponding Wilms tumors characterized for 11p15 genetic/epigenetic alterations and insulin-like growth factor-II expression.
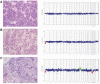
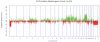
Results: The genomic profiles of PLNRs could be subdivided into three categories: those with no copy number changes (22 of 50, 44%); those with single, whole chromosome alterations (8 of 50, 16%); and those with multiple gains/losses (20 of 50, 40%). The most frequent aberrations included 1p- (7 of 50, 14%) +18 (6 of 50, 12%), +13 (5 of 50, 10%), and +12 (3 of 50, 6%). For the majority (19 of 25, 76%) of cases, the rest harbored a subset of the copy number changes in the associated Wilms tumor. We identified a temporal order of genomic changes, which occur during the insulin-like growth factor-II/PLNR pathway of Wilms tumorigenesis, with large-scale chromosomal alterations such as 1p-, +12, +13, and +18 regarded as "early" events. In some of the cases (24%), the PLNRs harbored large-scale copy number changes not observed in the concurrent Wilms tumor, including +10p, +14q, and +18.
Conclusions: These data suggest that although the evidence for PLNRs as precursors is compelling, not all lesions must necessarily undergo malignant transformation.
Figures



References
-
- Schedl A. Renal abnormalities and their developmental origin. Nat Rev Genet. 2007;8:791–802. - PubMed
-
- Rivera MN, Haber DA. Wilms' tumour: connecting tumorigenesis and organ development in the kidney. Nat Rev Cancer. 2005;5:699–712. - PubMed
-
- Beckwith JB, Kiviat NB, Bonadio JF. Nephrogenic rests, nephroblastomatosis, and the pathogenesis of Wilms' tumor. Pediatr Pathol. 1990;10:1–36. - PubMed
-
- Fukuzawa R, Reeve AE. Molecular pathology and epidemiology of nephrogenic rests and wilms tumors. J Pediatr Hematol Oncol. 2007;29:589–94. - PubMed
-
- Park S, Bernard A, Bove KE, et al. Inactivation of WT1 in nephrogenic rests, genetic precursors to Wilms' tumour. Nat Genet. 1993;5:363–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

