Characterization of the Arabidopsis nitrate transporter NRT1.6 reveals a role of nitrate in early embryo development
- PMID: 19050168
- PMCID: PMC2630450
- DOI: 10.1105/tpc.107.056788
Characterization of the Arabidopsis nitrate transporter NRT1.6 reveals a role of nitrate in early embryo development
Abstract
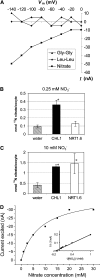
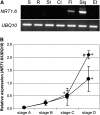
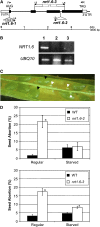
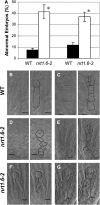
This study of the Arabidopsis thaliana nitrate transporter NRT1.6 indicated that nitrate is important for early embryo development. Functional analysis of cDNA-injected Xenopus laevis oocytes showed that NRT1.6 is a low-affinity nitrate transporter and does not transport dipeptides. RT-PCR, in situ hybridization, and beta-glucuronidase reporter gene analysis showed that expression of NRT1.6 is only detectable in reproductive tissue (the vascular tissue of the silique and funiculus) and that expression increases immediately after pollination, suggesting that NRT1.6 is involved in delivering nitrate from maternal tissue to the developing embryo. In nrt1.6 mutants, the amount of nitrate accumulated in mature seeds was reduced and the seed abortion rate increased. In the mutants, abnormalities (i.e., excessive cell division and loss of turgidity), were found mainly in the suspensor cells at the one- or two-cell stages of embryo development. The phenotype of the nrt1.6 mutants revealed a novel role of nitrate in early embryo development. Interestingly, the seed abortion rate of the mutant was reduced when grown under N-deficient conditions, suggesting that nitrate requirements in early embryo development can be modulated in response to external nitrogen changes.
Figures

References
-
- Alboresi, A., Gestin, C., Leydecker, M.T., Bedu, M., Meyer, C., and Truong, H.N. (2005). Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant Cell Environ. 28 500–512. - PubMed
-
- Chiang, C.-S., Stacey, G., and Tsay, Y.-F. (2004). Mechanisms and functional properties of two peptide transporters, AtPTR2 and fPTR2. J. Biol. Chem. 279 30150–30157. - PubMed
-
- Chiu, C.C., Lin, C.S., Hsia, A.P., Su, R.C., Lin, H.L., and Tsay, Y.F. (2004). Mutation of a nitrate transporter, AtNRT1:4, results in a reduced petiole nitrate content and altered leaf development. Plant Cell Physiol. 45 1139–1148. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

