In-frame deletion in the seventh immunoglobulin-like repeat of filamin C in a family with myofibrillar myopathy
- PMID: 19050726
- PMCID: PMC2672961
- DOI: 10.1038/ejhg.2008.226
In-frame deletion in the seventh immunoglobulin-like repeat of filamin C in a family with myofibrillar myopathy
Abstract
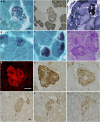
Myofibrillar myopathies (MFMs) are an expanding and increasingly recognized group of neuromuscular disorders caused by mutations in DES, CRYAB, MYOT, and ZASP. The latest gene to be associated with MFM was FLNC; a p.W2710X mutation in the 24th immunoglobulin-like repeat of filamin C was shown to be the cause of a distinct type of MFM in several German families. We studied an International cohort of 46 patients from 39 families with clinically and myopathologically confirmed MFM, in which DES, CRYAB, MYOT, and ZASP mutations have been excluded. In patients from an unrelated family a 12-nucleotide deletion (c.2997_3008del) in FLNC resulting in a predicted in-frame four-residue deletion (p.Val930_Thr933del) in the seventh repeat of filamin C was identified. Both affected family members, mother and daughter, but not unrelated control individuals, carried the p.Val930_Thr933del mutation. The mutation is transcribed and, based on myopathological features and immunoblot analysis, it leads to an accumulation of dysfunctional filamin C in the myocytes. The study results suggest that the novel p.Val930_Thr933del mutation in filamin C is the cause of MFM but also indicate that filamin C mutations are a comparatively rare cause of MFM.
Figures


References
-
- Nakano S, Engel AG, Waclawik AJ, et al. Myofibrillar myopathy with abnormal foci of desmin positivity. 1. Light and electron microscopy analysis of 10 cases. J Neuropathol Exp Neurol. 1996:55, 549–562. - PubMed
-
- Engel AG. Myofibrillar myopathy. Ann Neurol. 1999;46:681–683. - PubMed
-
- Selcen D, Ohno K, Engel AG. Myofibrillar myopathy: clinical, morphological and genetic studies in 63 patients. Brain. 2004;127:439–451. - PubMed
-
- Vicart P, Caron A, Guicheney P, et al. A missense mutation in the alphaB-crystallin chaperone gene causes a desmin-related myopathy. Nat Genet. 1998;20:92–95. - PubMed
-
- Selcen D, Engel AG.Myofibrillar myopathy GeneReviews , ( http://www.genereviews.org/ ) 2008
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

