Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection
- PMID: 19052094
- PMCID: PMC2643775
- DOI: 10.1128/JVI.01888-08
Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection
Abstract
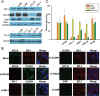
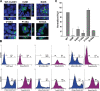
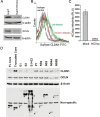
A tight junction (TJ) protein, claudin-1 (CLDN1), was identified recently as a key factor for hepatitis C virus (HCV) entry. Here, we show that another TJ protein, occludin, is also required for HCV entry. Mutational study of CLDN1 revealed that its tight junctional distribution plays an important role in mediating viral entry. Together, these data support the model in which HCV enters liver cells from the TJ. Interestingly, HCV infection of Huh-7 hepatoma cells downregulated the expression of CLDN1 and occludin, preventing superinfection. The altered TJ protein expression may contribute to the morphological and functional changes observed in HCV-infected hepatocytes.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

