Formin proteins of the DAAM subfamily play a role during axon growth
- PMID: 19052223
- PMCID: PMC6671601
- DOI: 10.1523/JNEUROSCI.2727-08.2008
Formin proteins of the DAAM subfamily play a role during axon growth
Abstract
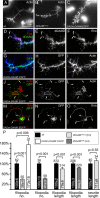
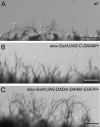
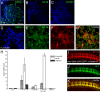
The regulation of growth cone actin dynamics is a critical aspect of axonal growth control. Among the proteins that are directly involved in the regulation of actin dynamics, actin nucleation factors play a pivotal role by promoting the formation of novel actin filaments. However, the essential nucleation factors in developing neurons have so far not been clearly identified. Here, we show expression data, and use true loss-of-function analysis and targeted expression of activated constructs to demonstrate that the Drosophila formin DAAM plays a critical role in axonal morphogenesis. In agreement with this finding, we show that dDAAM is required for filopodia formation at axonal growth cones. Our genetic interaction, immunoprecipitation and protein localization studies argue that dDAAM acts in concert with Rac GTPases, Profilin and Enabled during axonal growth regulation. We also show that mouse Daam1 rescues the CNS defects observed in dDAAM mutant flies to a high degree, and vice versa, that Drosophila DAAM induces the formation of neurite-like protrusions when expressed in mouse P19 cells, strongly suggesting that the function of DAAM in developing neurons has been conserved during evolution.
Figures




References
-
- Aspenström P, Richnau N, Johansson AS. The diaphanous-related formin DAAM1 collaborates with the Rho GTPases RhoA and Cdc42, CIP4 and Src in regulating cell morphogenesis and actin dynamics. Exp Cell Res. 2006;312:2180–2194. - PubMed
-
- Bachmann C, Fischer L, Walter U, Reinhard M. The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J Biol Chem. 1999;274:23549–23557. - PubMed
-
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, Gertler FB. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–521. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
