Characterizing heterogeneous cellular responses to perturbations
- PMID: 19052231
- PMCID: PMC2614757
- DOI: 10.1073/pnas.0807038105
Characterizing heterogeneous cellular responses to perturbations
Abstract
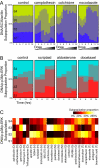
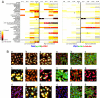
Cellular populations have been widely observed to respond heterogeneously to perturbation. However, interpreting the observed heterogeneity is an extremely challenging problem because of the complexity of possible cellular phenotypes, the large dimension of potential perturbations, and the lack of methods for separating meaningful biological information from noise. Here, we develop an image-based approach to characterize cellular phenotypes based on patterns of signaling marker colocalization. Heterogeneous cellular populations are characterized as mixtures of phenotypically distinct subpopulations, and responses to perturbations are summarized succinctly as probabilistic redistributions of these mixtures. We apply our method to characterize the heterogeneous responses of cancer cells to a panel of drugs. We find that cells treated with drugs of (dis-)similar mechanism exhibit (dis-)similar patterns of heterogeneity. Despite the observed phenotypic diversity of cells observed within our data, low-complexity models of heterogeneity were sufficient to distinguish most classes of drug mechanism. Our approach offers a computational framework for assessing the complexity of cellular heterogeneity, investigating the degree to which perturbations induce redistributions of a limited, but nontrivial, repertoire of underlying states and revealing functional significance contained within distinct patterns of heterogeneous responses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Bahar R, et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. - PubMed
-
- Bar-Even A, et al. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–643. - PubMed
-
- Colman-Lerner A, et al. Regulated cell-to-cell variation in a cell-fate decision system. Nature. 2005;437:699–706. - PubMed
-
- Ferrell JE, Jr, Machleder EM. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–898. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

