Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli
- PMID: 19052235
- PMCID: PMC2614783
- DOI: 10.1073/pnas.0807227105
Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli
Abstract
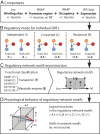
Broad-acting transcription factors (TFs) in bacteria form regulons. Here, we present a 4-step method to fully reconstruct the leucine-responsive protein (Lrp) regulon in Escherichia coli K-12 MG 1655 that regulates nitrogen metabolism. Step 1 is composed of obtaining high-resolution ChIP-chip data for Lrp, the RNA polymerase and expression profiles under multiple environmental conditions. We identified 138 unique and reproducible Lrp-binding regions and classified their binding state under different conditions. In the second step, the analysis of these data revealed 6 distinct regulatory modes for individual ORFs. In the third step, we used the functional assignment of the regulated ORFs to reconstruct 4 types of regulatory network motifs around the metabolites that are affected by the corresponding gene products. In the fourth step, we determined how leucine, as a signaling molecule, shifts the regulatory motifs for particular metabolites. The physiological structure that emerges shows the regulatory motifs for different amino acid fall into the traditional classification of amino acid families, thus elucidating the structure and physiological functions of the Lrp-regulon. The same procedure can be applied to other broad-acting TFs, opening the way to full bottom-up reconstruction of the transcriptional regulatory network in bacterial cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Yokoyama K, et al. Feast/famine regulatory proteins (FFRPs): Escherichia coli Lrp, AsnC and related archaeal transcription factors. FEMS Microbiol Rev. 2006;30:89–108. - PubMed
-
- Newman EB, Lin R. Leucine-responsive regulatory protein: a global regulator of gene expression in E. coli. Annu Rev Microbiol. 1995;49:747–775. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

