Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules
- PMID: 19052240
- PMCID: PMC2593616
- DOI: 10.1073/pnas.0810634105
Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules
Erratum in
- Proc Natl Acad Sci U S A. 2014 Feb 4;111(5):2046
Expression of concern in
-
Editorial expression of concern: Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules.Proc Natl Acad Sci U S A. 2014 Sep 9;111(36):13241. doi: 10.1073/pnas.1415649111. Epub 2014 Sep 2. Proc Natl Acad Sci U S A. 2014. PMID: 25197091 Free PMC article. No abstract available.
Abstract
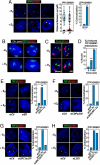
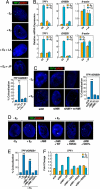
Although the role of liganded nuclear receptors in mediating coactivator/corepressor exchange is well-established, little is known about the potential regulation of chromosomal organization in the 3-dimensional space of the nucleus in achieving integrated transcriptional responses to diverse signaling events. Here, we report that ligand induces rapid interchromosomal interactions among specific subsets of estrogen receptor alpha-bound transcription units, with a dramatic reorganization of nuclear territories, which depends on the actions of nuclear actin/myosin-I machinery and dynein light chain 1. The histone lysine demethylase, LSD1, is required for these ligand-induced interactive loci to associate with distinct interchromatin granules, long thought to serve as "storage" sites for the splicing machinery, some critical transcription elongation factors, and various chromatin remodeling complexes. We demonstrate that this 2-step nuclear rearrangement is essential for achieving enhanced, coordinated transcription of nuclear receptor target genes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Dennis AP, O'Malley BW. Rush hour at the promoter: How the ubiquitin proteasome pathway polices the traffic flow of nuclear receptor-dependent transcription. J Steroid Biochem Mol Biol. 2005;93:139–151. - PubMed
-
- Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: A coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. - PubMed
-
- Carroll JS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. - PubMed
-
- Carroll JS, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HL088129/HL/NHLBI NIH HHS/United States
- R01 HL088129/HL/NHLBI NIH HHS/United States
- R33 CA114184/CA/NCI NIH HHS/United States
- DK018477/DK/NIDDK NIH HHS/United States
- R37 DK039949/DK/NIDDK NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- HL65445/HL/NHLBI NIH HHS/United States
- R01 CA052599/CA/NCI NIH HHS/United States
- R01 CA097134/CA/NCI NIH HHS/United States
- R01 DK018477/DK/NIDDK NIH HHS/United States
- NS034934/NS/NINDS NIH HHS/United States
- R01 DK039949/DK/NIDDK NIH HHS/United States
- CA52599/CA/NCI NIH HHS/United States
- GM049369/GM/NIGMS NIH HHS/United States
- CA97134/CA/NCI NIH HHS/United States
- R01 HL065445/HL/NHLBI NIH HHS/United States
- DK39949/DK/NIDDK NIH HHS/United States
- CA114184/CA/NCI NIH HHS/United States
- R01 GM049369/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

