A bioinformatician's guide to metagenomics
- PMID: 19052320
- PMCID: PMC2593568
- DOI: 10.1128/MMBR.00009-08
A bioinformatician's guide to metagenomics
Abstract
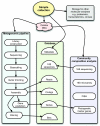
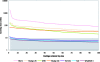
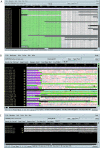
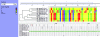
As random shotgun metagenomic projects proliferate and become the dominant source of publicly available sequence data, procedures for the best practices in their execution and analysis become increasingly important. Based on our experience at the Joint Genome Institute, we describe the chain of decisions accompanying a metagenomic project from the viewpoint of the bioinformatic analysis step by step. We guide the reader through a standard workflow for a metagenomic project beginning with presequencing considerations such as community composition and sequence data type that will greatly influence downstream analyses. We proceed with recommendations for sampling and data generation including sample and metadata collection, community profiling, construction of shotgun libraries, and sequencing strategies. We then discuss the application of generic sequence processing steps (read preprocessing, assembly, and gene prediction and annotation) to metagenomic data sets in contrast to genome projects. Different types of data analyses particular to metagenomes are then presented, including binning, dominant population analysis, and gene-centric analysis. Finally, data management issues are presented and discussed. We hope that this review will assist bioinformaticians and biologists in making better-informed decisions on their journey during a metagenomic project.
Figures



References
-
- Abe, T., H. Sugawara, M. Kinouchi, S. Kanaya, and T. Ikemura. 2005. Novel phylogenetic studies of genomic sequence fragments derived from uncultured microbe mixtures in environmental and clinical samples. DNA Res. 12281-290. - PubMed
-
- Achtman, M., and M. Wagner. 2008. Microbial diversity and the genetic nature of microbial species. Nat. Rev. Microbiol. 6431-440. - PubMed
-
- Allen, E. E., and J. F. Banfield. 2005. Community genomics in microbial ecology and evolution. Nat. Rev. Microbiol. 3489-498. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

