Mechanisms of severe acute respiratory syndrome pathogenesis and innate immunomodulation
- PMID: 19052324
- PMCID: PMC2593566
- DOI: 10.1128/MMBR.00015-08
Mechanisms of severe acute respiratory syndrome pathogenesis and innate immunomodulation
Abstract
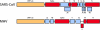
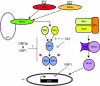
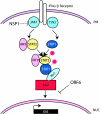
The modulation of the immune response is a common practice of many highly pathogenic viruses. The emergence of the highly pathogenic coronavirus severe acute respiratory virus (SARS-CoV) serves as a robust model system to elucidate the virus-host interactions that mediate severe end-stage lung disease in humans and animals. Coronaviruses encode the largest positive-sense RNA genome of approximately 30 kb, encode a variety of replicase and accessory open reading frames that are structurally unique, and encode novel enzymatic functions among RNA viruses. These viruses have broad or specific host ranges, suggesting the possibility of novel strategies for targeting and regulating host innate immune responses following virus infection. Using SARS-CoV as a model, we review the current literature on the ability of coronaviruses to interact with and modify the host intracellular environment during infection. These studies are revealing a rich set of novel viral proteins that engage, modify, and/or disrupt host cell signaling and nuclear import machinery for the benefit of virus replication.
Figures
References
-
- Akira, S., and H. Hemmi. 2003. Recognition of pathogen-associated molecular patterns by TLR family. Immunol. Lett. 8585-95. - PubMed
-
- Almazan, F., M. L. Dediego, C. Galan, D. Escors, E. Alvarez, J. Ortego, I. Sola, S. Zuniga, S. Alonso, J. L. Moreno, A. Nogales, C. Capiscol, and L. Enjuanes. 2006. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 8010900-10906. - PMC - PubMed
-
- Ank, N., H. West, and S. R. Paludan. 2006. IFN-lambda: novel antiviral cytokines. J. Interf. Cytok. Res. 26373-379. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

