Modeling a snap-action, variable-delay switch controlling extrinsic cell death
- PMID: 19053173
- PMCID: PMC2592357
- DOI: 10.1371/journal.pbio.0060299
Modeling a snap-action, variable-delay switch controlling extrinsic cell death
Abstract
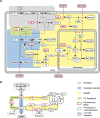
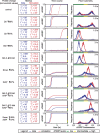
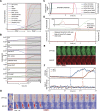
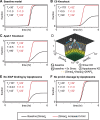
When exposed to tumor necrosis factor (TNF) or TNF-related apoptosis-inducing ligand (TRAIL), a closely related death ligand and investigational therapeutic, cells enter a protracted period of variable duration in which only upstream initiator caspases are active. A subsequent and sudden transition marks activation of the downstream effector caspases that rapidly dismantle the cell. Thus, extrinsic apoptosis is controlled by an unusual variable-delay, snap-action switch that enforces an unambiguous choice between life and death. To understand how the extrinsic apoptosis switch functions in quantitative terms, we constructed a mathematical model based on a mass-action representation of known reaction pathways. The model was trained against experimental data obtained by live-cell imaging, flow cytometry, and immunoblotting of cells perturbed by protein depletion and overexpression. The trained model accurately reproduces the behavior of normal and perturbed cells exposed to TRAIL, making it possible to study switching mechanisms in detail. Model analysis shows, and experiments confirm, that the duration of the delay prior to effector caspase activation is determined by initiator caspase-8 activity and the rates of other reactions lying immediately downstream of the TRAIL receptor. Sudden activation of effector caspases is achieved downstream by reactions involved in permeabilization of the mitochondrial membrane and relocalization of proteins such as Smac. We find that the pattern of interactions among Bcl-2 family members, the partitioning of Smac from its binding partner XIAP, and the mechanics of pore assembly are all critical for snap-action control.
Conflict of interest statement
Figures






References
-
- Green DR, Evan GI. A matter of life and death. Cancer Cell. 2002;1:19–30. - PubMed
-
- Wang J, Zheng L, Lobito A, Chan FK, Dale J, et al. Inherited human Caspase 10 mutations underlie defective lymphocyte and dendritic cell apoptosis in autoimmune lymphoproliferative syndrome type II. Cell. 1999;98:47–58. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

