MFS transportome of the human pathogenic yeast Candida albicans
- PMID: 19055746
- PMCID: PMC2636803
- DOI: 10.1186/1471-2164-9-579
MFS transportome of the human pathogenic yeast Candida albicans
Abstract
Background: The major facilitator superfamily (MFS) is one of the two largest superfamilies of membrane transporters present ubiquitously in bacteria, archaea, and eukarya and includes members that function as uniporters, symporters or antiporters. We report here the complete transportome of MFS proteins of a human pathogenic yeast Candida albicans.
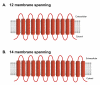
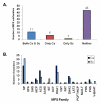
Results: Computational analysis of C. albicans genome enabled us to identify 95 potential MFS proteins which clustered into 17 families using Saier's Transport Commission (TC) system. Among these SP, DHA1, DHA2 and ACS represented major families consisting of 22, 22, 9 and 16 members, respectively. Family designations in C. albicans were validated by subjecting Saccharomyces cerevisiae genome to TC system. Based on the published available genomics/proteomics data, 87 of the putative MFS genes of C. albicans were found to express either at mRNA or protein levels. We checked the expression of the remaining 8 genes by using RT-PCR and observed that they are not expressed under basal growth conditions implying that either these 8 genes are expressed under specific growth conditions or they may be candidates for pseudogenes.
Conclusion: The in silico characterisation of MFS transporters in Candida albicans genome revealed a large complement of MFS transporters with most of them showing expression. Considering the clinical relevance of C. albicans and role of MFS members in antifungal resistance and nutrient transport, this analysis would pave way for identifying their physiological relevance.
Figures
References
-
- Pasrija R, Krishnamurthy S, Prasad T, Ernst JF, Prasad R. Squalene epoxidase encoded by ERG1 affects morphogenesis and drug susceptibilities of Candida albicans. J Antimicrob Chemother. 2005;55:905–913. - PubMed
-
- Prasad R, Gaur NA, Gaur M, Komath SS. Efflux pumps in drug resistance of Candida. Infect Disord Drug Targets. 2006;6:69–83. - PubMed
-
- Lopez-Ribot JL, McAtee RK, Lee LN, Kirkpatrick WR, White TC, Sanglard D, Patterson TF. Distinct patterns of gene expression associated with development of fluconazole resistance in serial Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob Agents Chemother. 1998;42:2932–2937. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

