Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol
- PMID: 19055772
- PMCID: PMC2621116
- DOI: 10.1186/1475-2859-7-36
Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol
Abstract
Background: Increasing energy costs and environmental concerns have motivated engineering microbes for the production of "second generation" biofuels that have better properties than ethanol.
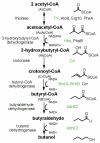
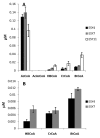
Results and conclusion: Saccharomyces cerevisiae was engineered with an n-butanol biosynthetic pathway, in which isozymes from a number of different organisms (S. cerevisiae, Escherichia coli, Clostridium beijerinckii, and Ralstonia eutropha) were substituted for the Clostridial enzymes and their effect on n-butanol production was compared. By choosing the appropriate isozymes, we were able to improve production of n-butanol ten-fold to 2.5 mg/L. The most productive strains harbored the C. beijerinckii 3-hydroxybutyryl-CoA dehydrogenase, which uses NADH as a co-factor, rather than the R. eutropha isozyme, which uses NADPH, and the acetoacetyl-CoA transferase from S. cerevisiae or E. coli rather than that from R. eutropha. Surprisingly, expression of the genes encoding the butyryl-CoA dehydrogenase from C. beijerinckii (bcd and etfAB) did not improve butanol production significantly as previously reported in E. coli. Using metabolite analysis, we were able to determine which steps in the n-butanol biosynthetic pathway were the most problematic and ripe for future improvement.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

