Integrated multi-level quality control for proteomic profiling studies using mass spectrometry
- PMID: 19055809
- PMCID: PMC2657802
- DOI: 10.1186/1471-2105-9-519
Integrated multi-level quality control for proteomic profiling studies using mass spectrometry
Abstract
Background: Proteomic profiling using mass spectrometry (MS) is one of the most promising methods for the analysis of complex biological samples such as urine, serum and tissue for biomarker discovery. Such experiments are often conducted using MALDI-TOF (matrix-assisted laser desorption/ionisation time-of-flight) and SELDI-TOF (surface-enhanced laser desorption/ionisation time-of-flight) MS. Using such profiling methods it is possible to identify changes in protein expression that differentiate disease states and individual proteins or patterns that may be useful as potential biomarkers. However, the incorporation of quality control (QC) processes that allow the identification of low quality spectra reliably and hence allow the removal of such data before further analysis is often overlooked. In this paper we describe rigorous methods for the assessment of quality of spectral data. These procedures are presented in a user-friendly, web-based program. The data obtained post-QC is then examined using variance components analysis to quantify the amount of variance due to some of the factors in the experimental design.
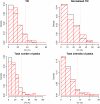
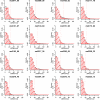
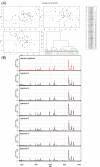
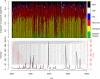
Results: Using data from a SELDI profiling study of serum from patients with different levels of renal function, we show how the algorithms described in this paper may be used to detect systematic variability within and between sample replicates, pooled samples and SELDI chips and spots. Manual inspection of those spectral data that were identified as being of poor quality confirmed the efficacy of the algorithms. Variance components analysis demonstrated the relatively small amount of technical variance attributable to day of profile generation and experimental array.
Conclusion: Using the techniques described in this paper it is possible to reliably detect poor quality data within proteomic profiling experiments undertaken by MS. The removal of these spectra at the initial stages of the analysis substantially improves the confidence of putative biomarker identification and allows inter-experimental comparisons to be carried out with greater confidence.
Figures


References
-
- Ricolleau G, Charbonnel C, Lode L, Loussouarn D, Joalland MP, Bogumil R, Jourdain S, Minvielle S, Campone M, Deporte-Fety R, Campion L, Jezequel P. Surface-enhanced laser desorption/ionization time of flight mass spectrometry protein profiling identifies ubiquitin and ferritin light chain as prognostic biomarkers in node-negative breast cancer tumors. Proteomics. 2006;6:1963–1975. doi: 10.1002/pmic.200500283. - DOI - PubMed
-
- Taguchi F, Solomon B, Gregorc V, Roder H, Gray R, Kasahara K, Nishio M, Brahmer J, Spreafico A, Ludovini V, Massion PP, Dziadziuszko R, Schiller J, Grigorieva J, Tsypin M, Hunsucker SW, Caprioli R, Duncan MW, Hirsch FR, Bunn PA, Carbone DP. Mass spectrometry to classify non-small-cell lung cancer patients for clinical outcome after treatment with epidermal growth factor receptor tyrosine kinase inhibitors: A multicohort cross-institutional study. Journal of the National Cancer Institute. 2007;99:838–846. doi: 10.1093/jnci/djk195. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

