Exploring the role of the immune response in preventing antibiotic resistance
- PMID: 19056402
- PMCID: PMC5814249
- DOI: 10.1016/j.jtbi.2008.10.025
Exploring the role of the immune response in preventing antibiotic resistance
Abstract
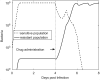
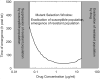
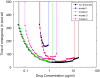
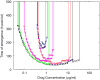
For many bacterial infections, drug resistant mutants are likely present by the time antibiotic treatment starts. Nevertheless, such infections are often successfully cleared. It is commonly assumed that this is due to the combined action of drug and immune response, the latter facilitating clearance of the resistant population. However, most studies of drug resistance emergence during antibiotic treatment focus almost exclusively on the dynamics of bacteria and the drug and neglect the contribution of immune defenses. Here, we develop and analyze several mathematical models that explicitly include an immune response. We consider different types of immune responses and investigate how each impacts the emergence of resistance. We show that an immune response that retains its strength despite a strong drug-induced decline of bacteria numbers considerably reduces the emergence of resistance, narrows the mutant selection window, and mitigates the effects of non-adherence to treatment. Additionally, we show that compared to an immune response that kills bacteria at a constant rate, one that trades reduced killing at high bacterial load for increased killing at low bacterial load is sometimes preferable. We discuss the predictions and hypotheses derived from this study and how they can be tested experimentally.
Figures


References
-
- Ambrose PG, Bhavnani SM, Rubino CM, Louie A, Gumbo T, Forrest A, Drusano GL. Pharmacokinetics–pharmacodynamics of antimicrobial therapy: it's not just for mice anymore. Clin Infect Dis. 2007;44(1):79–86. URL 〈 http://dx.doi.org/10.1086/510079〉. - DOI - PubMed
-
- Andes D, Craig WA. Animal model pharmacokinetics and pharmacody-namics: a critical review. Int J Antimicrob Agents. 2002;19(4):261–268. - PubMed
-
- Antia R, Bergstrom CT, Pilyugin SS, Kaech SM, Ahmed R. Models of CD8+ responses: 1. What is the antigen-independent proliferation program. J Theor Biol. 2003;221(4):585–598. - PubMed
-
- Austin DJ, White NJ, Anderson RM. The dynamics of drug action on the within-host population growth of infectious agents: melding pharmacokinetics with pathogen population dynamics. J Theor Biol. 1998;194(3):313–339. URL 〈 http://dx.doi.org/10.1006/jtbi.1997.0438〉. - DOI - PubMed
-
- Bajenoff M, Wurtz O, Guerder S. Repeated antigen exposure is necessary for the differentiation, but not the initial proliferation, of naive CD4(+) T cells. J Immunol. 2002;168(4):1723–1729. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

