Structure and function of colicin S4, a colicin with a duplicated receptor-binding domain
- PMID: 19056731
- PMCID: PMC2649078
- DOI: 10.1074/jbc.M808504200
Structure and function of colicin S4, a colicin with a duplicated receptor-binding domain
Abstract
Colicins are plasmid-encoded toxic proteins produced by Escherichia coli strains to kill other E. coli strains that lack the corresponding immunity protein. Colicins intrude into the host cell by exploiting existing transport, diffusion, or efflux systems. We have traced the way colicin S4 takes to execute its function and show that it interacts specifically with OmpW, OmpF, and the Tol system before it inserts its pore-forming domain into the cytoplasmic membrane. The common structural architecture of colicins comprises a translocation, a receptor-binding, and an activity domain. We have solved the crystal structure of colicin S4 to a resolution of 2.5 A, which shows a remarkably compact domain arrangement of four independent domains, including a unique domain duplication of the receptor-binding domain. Finally, we have determined the residues responsible for binding to the receptor OmpW by mutating exposed charged residues in one or both receptor-binding domains.
Figures



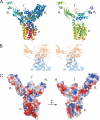

References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

