Biotin sensing at the molecular level
- PMID: 19056812
- PMCID: PMC2646212
- DOI: 10.3945/jn.108.095760
Biotin sensing at the molecular level
Abstract
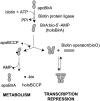
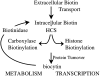
Biotin influences transcription in organisms from bacteria to humans. The enzyme, biotin protein ligase, which catalyzes post-transcriptional biotin addition to biotin-dependent carboxylases, plays a central roll in transmitting the demand for biotin to gene expression. The molecular mechanism of this communication in bacteria is well understood and involves competing protein:protein interactions. Biochemical measurements indicate that this competition is kinetically controlled. In humans, the biochemistry of biotin sensing at the transcriptional level is not well characterized. However, the biotin holoenzyme ligase (holocarboxylase synthetase) is proposed to both catalyze biotin addition to carboxylases and to histones in its metabolic and transcriptional roles, respectively. Control of human holocarboxylase synthetase function is, however, considerably more complex than the simple competitive protein protein interactions observed in bacterial systems.
Figures

References
-
- Norman AW. Minireview: vitamin D receptor: new assignments for an already busy receptor. Endocrinology. 2006;147:5542–8. - PubMed
-
- Weickert MJ, Adhya S. A family of bacterial regulators homologous to Gal and Lac repressors. J Biol Chem. 1992;267:15869–74. - PubMed
-
- Pennella MA, Giedroc DP. Structural determinants of metal selectivity in prokaryotic metal-responsive transcriptional regulators. Biometals. 2005;18:413–28. - PubMed
-
- Shi Y. Metabolic enzymes and coenzymes in transcription–a direct link between metabolism and transcription? Trends Genet. 2004;20:445–52. - PubMed
-
- Hall DA, Zhu H, Zhu X, Royce T, Gerstein M, Snyder M. Regulation of gene expression by a metabolic enzyme. Science. 2004;306:482–4. - PubMed

