Novel mutations in Moloney Murine Leukemia Virus reverse transcriptase increase thermostability through tighter binding to template-primer
- PMID: 19056821
- PMCID: PMC2632894
- DOI: 10.1093/nar/gkn952
Novel mutations in Moloney Murine Leukemia Virus reverse transcriptase increase thermostability through tighter binding to template-primer
Abstract
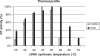
In an effort to increase the thermostability of Moloney Murine Leukemia Virus reverse transcriptase (MMLV RT), we screened random and site-saturation libraries for variants that show increased resistance to thermal inactivation. We discovered five mutations E69K, E302R, W313F, L435G and N454K that collectively increase the half-life of MMLV RT at 55 degrees C from less than 5 min to approximately 30 min in the presence of template-primer. In addition, these mutations alter the thermal profile by increasing specific activity of the pentuple mutant (M5) over a broad range of cDNA synthesis temperatures (25-70 degrees C). We further show that M5 generates higher cDNA yields and exhibits better RT-PCR performance compared to wild-type RT when used at high temperature to amplify RNA targets containing secondary structure. Finally, we demonstrate that M5 exhibits tighter binding (lower K(m)) to template-primer, which likely protects against heat inactivation.
Figures



References
-
- Molling K, Bolognesi DP, Bauer H, Busen W, Plassmann HW, Hausen P. Association of viral reverse transcriptase with an enzyme degrading the RNA moiety of RNA-DNA hybrids. Nat. New Biol. 1971;234:240–243. - PubMed
-
- Berger SL, Wallace DM, Puskas RS, Eschenfeldt WH. Reverse transcriptase and its associated ribonuclease H: interplay of two enzyme activities controls the yield of single-stranded complementary deoxyribonucleic acid. Biochemistry. 1983;22:2365–2372. - PubMed
-
- Krug MS, Berger SL. First-strand cDNA synthesis primed with oligo(dT) Methods Enzymol. 1987;152:316–325. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

