Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters
- PMID: 19056941
- PMCID: PMC2833333
- DOI: 10.1126/science.1162228
Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters
Abstract
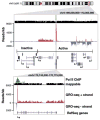
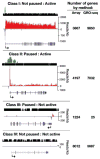
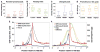
RNA polymerases are highly regulated molecular machines. We present a method (global run-on sequencing, GRO-seq) that maps the position, amount, and orientation of transcriptionally engaged RNA polymerases genome-wide. In this method, nuclear run-on RNA molecules are subjected to large-scale parallel sequencing and mapped to the genome. We show that peaks of promoter-proximal polymerase reside on approximately 30% of human genes, transcription extends beyond pre-messenger RNA 3' cleavage, and antisense transcription is prevalent. Additionally, most promoters have an engaged polymerase upstream and in an orientation opposite to the annotated gene. This divergent polymerase is associated with active genes but does not elongate effectively beyond the promoter. These results imply that the interplay between polymerases and regulators over broad promoter regions dictates the orientation and efficiency of productive transcription.
Figures

Comment in
-
Transcription. Gene expression--where to start?Science. 2008 Dec 19;322(5909):1804-5. doi: 10.1126/science.1168805. Science. 2008. PMID: 19095933 Free PMC article. No abstract available.
References
-
- Wold B, Myers RM. Nat Methods. 2008;5:19. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

