GeneDistiller--distilling candidate genes from linkage intervals
- PMID: 19057649
- PMCID: PMC2587712
- DOI: 10.1371/journal.pone.0003874
GeneDistiller--distilling candidate genes from linkage intervals
Abstract
Background: Linkage studies often yield intervals containing several hundred positional candidate genes. Different manual or automatic approaches exist for the determination of the gene most likely to cause the disease. While the manual search is very flexible and takes advantage of the researchers' background knowledge and intuition, it may be very cumbersome to collect and study the relevant data. Automatic solutions on the other hand usually focus on certain models, remain "black boxes" and do not offer the same degree of flexibility.
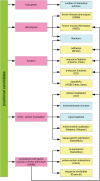
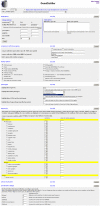
Methodology: We have developed a web-based application that combines the advantages of both approaches. Information from various data sources such as gene-phenotype associations, gene expression patterns and protein-protein interactions was integrated into a central database. Researchers can select which information for the genes within a candidate interval or for single genes shall be displayed. Genes can also interactively be filtered, sorted and prioritised according to criteria derived from the background knowledge and preconception of the disease under scrutiny.
Conclusions: GeneDistiller provides knowledge-driven, fully interactive and intuitive access to multiple data sources. It displays maximum relevant information, while saving the user from drowning in the flood of data. A typical query takes less than two seconds, thus allowing an interactive and explorative approach to the hunt for the candidate gene.
Access: GeneDistiller can be freely accessed at http://www.genedistiller.org.
Conflict of interest statement
Figures



References
-
- Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. - PubMed
-
- Schuelke M, Wagner KR, Stolz LE, Hubner C, Riebel T, et al. Myostatin mutation associated with gross muscle hypertrophy in a child. N Engl J Med. 2004;350:2682–2688. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

