APOBEC3G inhibits elongation of HIV-1 reverse transcripts
- PMID: 19057663
- PMCID: PMC2584787
- DOI: 10.1371/journal.ppat.1000231
APOBEC3G inhibits elongation of HIV-1 reverse transcripts
Abstract
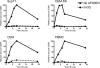
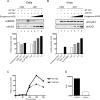
APOBEC3G (A3G) is a host cytidine deaminase that, in the absence of Vif, restricts HIV-1 replication and reduces the amount of viral DNA that accumulates in cells. Initial studies determined that A3G induces extensive mutation of nascent HIV-1 cDNA during reverse transcription. It has been proposed that this triggers the degradation of the viral DNA, but there is now mounting evidence that this mechanism may not be correct. Here, we use a natural endogenous reverse transcriptase assay to show that, in cell-free virus particles, A3G is able to inhibit HIV-1 cDNA accumulation not only in the absence of hypermutation but also without the apparent need for any target cell factors. We find that although reverse transcription initiates in the presence of A3G, elongation of the cDNA product is impeded. These data support the model that A3G reduces HIV-1 cDNA levels by inhibiting synthesis rather than by inducing degradation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

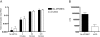

References
-
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. - PubMed
-
- Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol. 2008;26:317–353. - PubMed
-
- Holmes RK, Malim MH, Bishop KN. APOBEC-mediated viral restriction: not simply editing? Trends Biochem Sci. 2007;32:118–128. - PubMed
-
- Malim MH, Emerman M. HIV-1 accessory proteins—Ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3:388–398. - PubMed
-
- Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med. 2003;9:1404–1407. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

