The genomic analysis of lactic acidosis and acidosis response in human cancers
- PMID: 19057672
- PMCID: PMC2585811
- DOI: 10.1371/journal.pgen.1000293
The genomic analysis of lactic acidosis and acidosis response in human cancers
Abstract
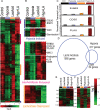
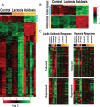
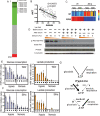
The tumor microenvironment has a significant impact on tumor development. Two important determinants in this environment are hypoxia and lactic acidosis. Although lactic acidosis has long been recognized as an important factor in cancer, relatively little is known about how cells respond to lactic acidosis and how that response relates to cancer phenotypes. We develop genome-scale gene expression studies to dissect transcriptional responses of primary human mammary epithelial cells to lactic acidosis and hypoxia in vitro and to explore how they are linked to clinical tumor phenotypes in vivo. The resulting experimental signatures of responses to lactic acidosis and hypoxia are evaluated in a heterogeneous set of breast cancer datasets. A strong lactic acidosis response signature identifies a subgroup of low-risk breast cancer patients having distinct metabolic profiles suggestive of a preference for aerobic respiration. The association of lactic acidosis response with good survival outcomes may relate to the role of lactic acidosis in directing energy generation toward aerobic respiration and utilization of other energy sources via inhibition of glycolysis. This "inhibition of glycolysis" phenotype in tumors is likely caused by the repression of glycolysis gene expression and Akt inhibition. Our study presents a genomic evaluation of the prognostic information of a lactic acidosis response independent of the hypoxic response. Our results identify causal roles of lactic acidosis in metabolic reprogramming, and the direct functional consequence of lactic acidosis pathway activity on cellular responses and tumor development. The study also demonstrates the utility of genomic analysis that maps expression-based findings from in vitro experiments to human samples to assess links to in vivo clinical phenotypes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Vaupel P. Tumor microenvironmental physiology and its implications for radiation oncology. Semin Radiat Oncol. 2004;14:198–206. - PubMed
-
- Helmlinger G, Yuan F, Dellian M, Jain RK. Interstitial pH and pO2 gradients in solid tumors in vivo: high-resolution measurements reveal a lack of correlation. Nat Med. 1997;3:177–182. - PubMed
-
- Vaupel P, Hockel M. Blood supply, oxygenation status and metabolic micromilieu of breast cancers: characterization and therapeutic relevance. Int J Oncol. 2000;17:869–879. - PubMed
-
- Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. - PubMed
-
- Cardone RA, Casavola V, Reshkin SJ. The role of disturbed pH dynamics and the Na+/H+ exchanger in metastasis. Nat Rev Cancer. 2005;5:786–795. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

