Cobalamin-dependent and cobamide-dependent methyltransferases
- PMID: 19059104
- PMCID: PMC2639622
- DOI: 10.1016/j.sbi.2008.11.005
Cobalamin-dependent and cobamide-dependent methyltransferases
Abstract
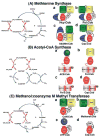
Methyltransferases that employ cobalamin cofactors, or their analogs the cobamides, as intermediates in catalysis of methyl transfer play vital roles in energy generation in anaerobic unicellular organisms. In a broader range of organisms they are involved in the conversion of homocysteine to methionine. Although the individual methyl transfer reactions catalyzed are simple S(N)2 displacements, the required change in coordination at the cobalt of the cobalamin or cobamide cofactors and the lability of the reduced Co(+1) intermediates introduces the necessity for complex conformational changes during the catalytic cycle. Recent spectroscopic and structural studies on several of these methyltransferases have helped to reveal the strategies by which these conformational changes are facilitated and controlled.
Figures



Similar articles
-
Structure-based perspectives on B12-dependent enzymes.Annu Rev Biochem. 1997;66:269-313. doi: 10.1146/annurev.biochem.66.1.269. Annu Rev Biochem. 1997. PMID: 9242908 Review.
-
Laboratory evolution of E. coli with a natural vitamin B12 analog reveals roles for cobamide uptake and adenosylation in methionine synthase-dependent growth.J Bacteriol. 2025 Feb 20;207(2):e0028424. doi: 10.1128/jb.00284-24. Epub 2025 Jan 28. J Bacteriol. 2025. PMID: 39873498 Free PMC article.
-
The reactivity of B12 cofactors: the proteins make a difference.Structure. 1996 May 15;4(5):505-12. doi: 10.1016/s0969-2126(96)00056-1. Structure. 1996. PMID: 8736549 Review.
-
Biochemistry of B12-cofactors in human metabolism.Subcell Biochem. 2012;56:323-46. doi: 10.1007/978-94-007-2199-9_17. Subcell Biochem. 2012. PMID: 22116707 Review.
-
Vitamin B12-derivatives-enzyme cofactors and ligands of proteins and nucleic acids.Chem Soc Rev. 2011 Aug;40(8):4346-63. doi: 10.1039/c1cs15118e. Epub 2011 Jun 20. Chem Soc Rev. 2011. PMID: 21687905 Review.
Cited by
-
Berberine-microbiota interplay: orchestrating gut health through modulation of the gut microbiota and metabolic transformation into bioactive metabolites.Front Pharmacol. 2023 Dec 7;14:1281090. doi: 10.3389/fphar.2023.1281090. eCollection 2023. Front Pharmacol. 2023. PMID: 38130410 Free PMC article.
-
Co+-H interaction inspired alternate coordination geometries of biologically important cob(I)alamin: possible structural and mechanistic consequences for methyltransferases.J Biol Inorg Chem. 2012 Oct;17(7):1107-21. doi: 10.1007/s00775-012-0924-x. Epub 2012 Aug 8. J Biol Inorg Chem. 2012. PMID: 22872137
-
Identification and characterization of a bacterial core methionine synthase.Sci Rep. 2020 Feb 7;10(1):2100. doi: 10.1038/s41598-020-58873-z. Sci Rep. 2020. PMID: 32034217 Free PMC article.
-
Metabolic engineering of Escherichia coli W3110 for the production of L-methionine.J Ind Microbiol Biotechnol. 2017 Jan;44(1):75-88. doi: 10.1007/s10295-016-1870-3. Epub 2016 Nov 14. J Ind Microbiol Biotechnol. 2017. PMID: 27844169
-
Antivitamins B12 -Some Inaugural Milestones.Chemistry. 2020 Dec 1;26(67):15438-15445. doi: 10.1002/chem.202003788. Epub 2020 Nov 3. Chemistry. 2020. PMID: 32956545 Free PMC article. Review.
References
-
- Matthews RG. Corrinoid- and cobalamin-dependent methyltransferases. In: Sigal A, Sigal H, Sigal RKO, editors. Metal-carbon bonds in enzymes and cofactors. Royal Society of Chemistry; 2009:in press. vol 6 of “Metal Ions in Life Sciences”.
-
- Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;438:90–93. - PubMed
-
- Goulding CW, Postigo D, Matthews RG. Cobalamin-dependent methionine synthase is a modular protein with distinct regions for binding homocysteine, methyltetrahydrofolate, cobalamin, and adenosylmethionine. Biochemistry. 1997;36:8082–8091. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

